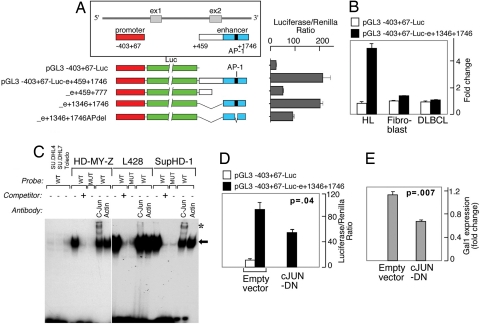
Fig. 2.
Gal1 transcription is regulated by an AP1-dependent enhancer. (A) Analysis of the AP1-dependent Gal1 enhancer. The previously described Gal1 promoter (21) and putative enhancer element including or lacking the predicted AP1-binding site (represented by a black bar) were cloned into a luciferase reporter vector, transiently transfected into cHL HD-MY-Z cells, and assayed for luciferase activities. Representative luciferase activities from three independent experiments are normalized to Renilla luciferase activity and are presented as bars ± SD. (B) Selective activity of the Gal1 enhancer. Classical HL, DLBCL, and fibroblast lines were transfected with either the Gal1 promoter-only vector (pGL3-Gal1−403 +67-Luc) or the promoter–enhancer construct (pGL3-Gal1403 +67-Luc-e1346–1746) and assessed as in A for their respective luciferase activities. (C) AP-1 dependence of the Gal1 enhancer in electrophoretic mobility shift assays. Nuclear extracts from DLBCL cell lines (DHL4, DHL7, and Toledo) or cHL cell lines (HD-MY-Z, L428, and SupHD1) were incubated with WT or mutant 32P-labeled, double-stranded DNA probe corresponding to AP1-binding site in Gal1 enhancer. Specific, unlabeled competitor and antibodies against c-Jun or β-actin (control) were included in certain assays as indicated. The gel-shift band corresponding to the probe–protein complex is indicated with an arrow, and supershift bands corresponding to the probe–protein–antibody complex are noted with asterisks. (D) c-Jun dependence of the Gal1 enhancer. HD-MY-Z cells were cotransfected with the Gal1 promotor-only vector or the Gal1 promotor–enhancer construct with either the dominant-negative c-Jun (c-Jun-DN) construct (c-Jun-DN) or empty vector. Luciferease activities were determined as in A. (E) AP1 inhibition decreases Gal1 transcript abundance. HD-MY-Z cells were transfected with either c-Jun-DN or empty vector. Thereafter, relative Gal1 mRNA abundance was assessed by real-time QPCR.