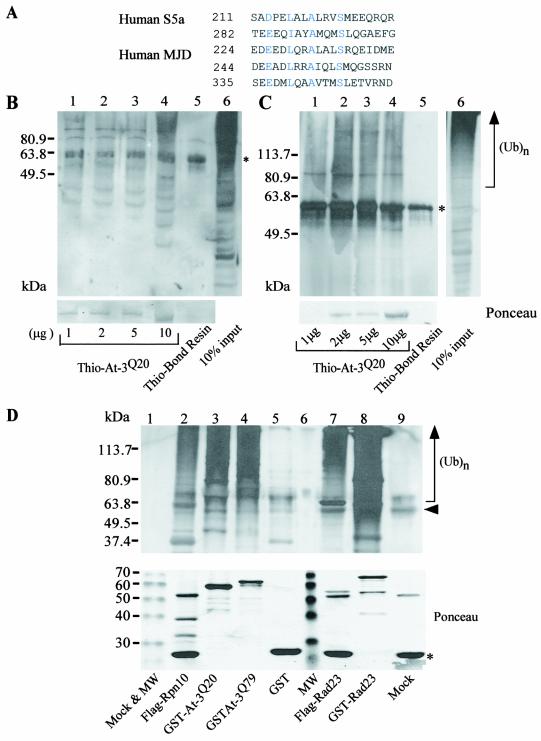
FIG. 1.
A sequence motif (UIM) in S5a that can bind ubiquitinated proteins is present in ataxin-3. (A) Alignment of conserved regions of S5a and ataxin-3. The position of the amino acid residue at the N terminus of the conserved UIM sequence is shown on the left. Amino acid residues in blue are identical or highly conserved. (B) Ataxin-3 can bind ubiquitinated proteins in yeast cell extracts. Protein extracts were prepared from a wild-type yeast strain, and 1 mg was incubated with various amounts of Thio-ataxin-3Q20 that was purified from E. coli. After extensive washing, the proteins that were bound to Thio-ataxin-3 were separated in an SDS-12% polyacrylamide gel, transferred to nitrocellulose filters (Bio-Rad), and incubated with ubiquitin-specific antibodies (Sigma). The immunoblot was developed with enhanced chemiluminescence (Dupont, NEN). Lanes 1 to 4 contain, respectively, 1, 2, 5, and 10 μg of Thio-ataxin-3Q20 on affinity beads. Lane 5, control in which Thio-Bond resin was incubated with a yeast cell extract. A sample of the yeast cell extract (10% input) is shown in lane 6. The positions of molecular size standards are shown in the left margin. Asterisks in panels B and C indicate a nonspecific band (from Thio-Bond resin) that cross-reacted with anti-ubiquitin antibodies. Ponceau staining shows the amount of ataxin-3 protein used in the pulldown assay. At-3, ataxin-3. (C) Ataxin-3 can bind ubiquitinated proteins from NT2 human cell extracts. Protein extracts were prepared from NT2 cells, applied to various amounts of Thio-ataxin-3Q20 immobilized onbeads, and examined as described for panel B, but in an SDS-10% polyacrylamide gel. Lanes are as in panel B, except that lane 6 corresponds to 10% input of NT2 cell extract. Ubiquitinated proteins are indicated by the designation (Ub)n in the right margin. Ponceau staining shows the amount of ataxin-3 protein used in the pulldown assay. (D) Ataxin-3 interacts with ubiquitinated proteins in vivo in yeast cells. Protein extracts were prepared from strains expressing FLAG-Rpn10, GST-ataxin-3Q20, GST-ataxin-3Q79, GST, FLAG-Rad23, or GST-Rad23 from the copper-inducible CUP1 promoter. Fusion proteins were isolated either by pulldown with glutathione Sepharose (Amersham Pharmacia Biotech) or by immunoprecipitation with FLAG-agarose (Sigma). The bound proteins were separated in an SDS-10% polyacrylamide gel and transferred to nitrocellulose membranes, and ubiquitinated proteins were detected. Lanes 1 and 9, mock reactions in which an extract derived from wild-type yeast that did not overexpress any of the fusion proteins was applied to either glutathione Sepharose or FLAG-agarose, respectively; lane 2, FLAG-Rpn10; lane 3, GST-ataxin-3Q20; lane 4, GST-ataxin-3Q79; lane 5, GST; lane 6, molecular weight (MW) markers; lane 7, FLAG-Rad23; lane 8, GST-Rad23. For lanes 2, 7, and 9, an arrowhead indicates the immunoglobulin heavy chain. Asterisk, immunoglobulin light chain detected as an ∼23-kDa band in lanes 2, 7, and 9 in the lower panel. Ponceau staining shows the purified proteins prior to incubation with anti-ubiquitin antibodies, with theoretical molecular sizes as follows: for Flag-Rpn10, ∼30 kDa; for GST-ataxin-3Q20, ∼69 kDa; for GST-ataxin-3Q79, ∼78 kDa; for GST, ∼28 kDa; for FLAG-Rad23, ∼43 kDa; and for GST-Rad23, ∼70 kDa. Note that apparent molecular sizes as determined from SDS-PAGE are slightly different from those determined theoretically.