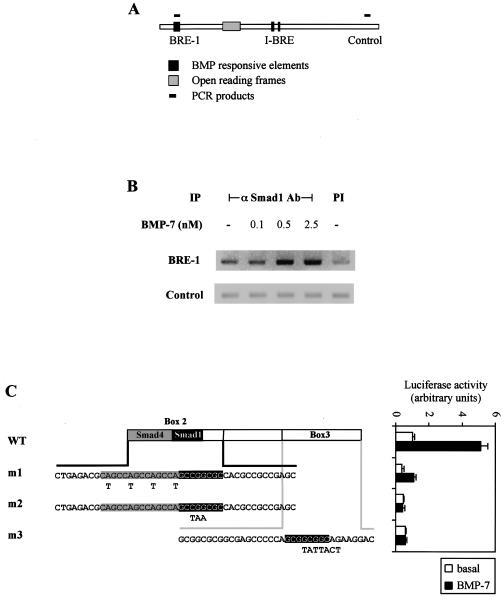
FIG. 3.
Smad1 binds to BRE-1 in living cells. (A) Schematic representation of PCR products amplified following ChIP assays. (B) ChIP of Smad1 bound to BRE-1. HepG2 cells were treated for 1 h with increasing concentrations of BMP-7. Protein-DNA complexes were cross-linked by formaldehyde treatment, cells were lysed, and DNA was sheared by sonication. Cell lysates were subjected to immunoprecipitation with an anti-Smad1 antibody. DNA recovered from the immunoprecipitation was amplified by PCR. PI, preimmune; −, absent. (C) Diagrammed clones derived from BRE-1 were transiently transfected in HepG2 cells with 0.03 μg of Smad1 and Smad4 cDNA/well. Cells were incubated overnight in the absence (white bars) or presence (black bars) of 5 nM BMP-7. The relative luciferase activities of cell lysates were determined. Mutations created in box 2 or box 3 are indicated in bold letters under the appropriate sequences. The putative Smad4 and Smad1 binding sites are highlighted in gray and black, respectively. WT, wild type.