Figure 2.
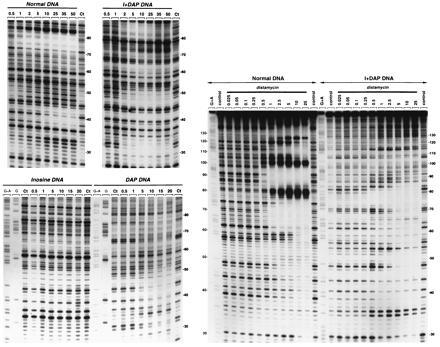
DNase I footprinting of mithramycin (Left) and distamycin (Right) on the Watson strand of tyrT(A93) DNA containing the four natural nucleotides (Normal DNA), inosine residues in place of guanosine (inosine DNA), DAP in place of adenine (DAP DNA) or inosine and DAP residues in place of guanosine and adenine respectively (I+DAP DNA). The products of DNase I digestion were identified by reference to the Maxam–Gilbert markers (lanes G and G+A), taking into account the difference in mobility of the fragments due to the presence or absence of a 3′-phosphate group. Control lanes (Ct) show the products resulting from limited DNase I digestion in the absence of ligand. The remaining lanes show the products of digestion in the presence of the indicated antibiotic concentrations (expressed as micromolar). Numbers at the side of the gels refer to the numbering scheme used in Fig. 3.
