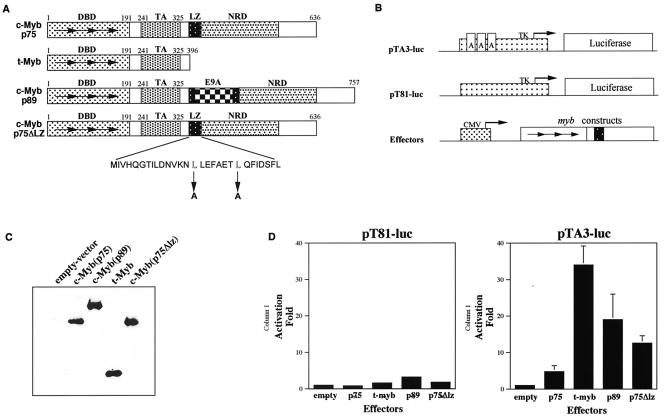
FIG. 1.
Transcriptional transactivation by p75 c-Myb, p75Δlz c-Myb, p89 c-Myb, and t-Myb. (A) Schematic representations of wild-type p75 c-Myb and p89 c-Myb and mutants p75Δlz c-Myb and t-Myb are depicted. The numbers above each diagram are positions of amino acid residues in each corresponding region. Horizontal arrows represent the three 51- to 52-amino-acid repeats that constitute the DNA-binding domain. The stretch of amino acids below represents the putative leucine zipper motif present in wild-type p75 c-Myb. The downward arrows indicate the two leucine to alanine mutations made in order to construct p75Δlz c-Myb. DBD, DNA binding domain; TA, transactivation domain; NRD, negative regulatory domain; LZ, leucine zipper; E9A, exon 9A. (B) Schematics of the reporter and effector plasmids used in transient transactivation assays. The dotted box represents the promoters, and the arrows in the promoters represent the starting sites of transcription. The arrows in Myb represent the three 51- to 52-amino-acid repeats of the DNA binding domain, and the black box represents the transactivation domain. The boxes labeled A in pTA3-luc represent the three Myb-binding sites. TK, herpes simplex virus thymidine kinase promoter; CMV, immediate-early promoter for cytomegalovirus. (C) Transient expression of effector molecules. Cell lysates from QT6 cells transfected with different myb expression plasmids and expressing equal amounts of firefly luciferase activity were subjected to Western blot analysis and probed with an antibody raised against c-Myb. (D) Transcriptional activation by Myb proteins. Each myb expression plasmid was transfected into QT6 cells with reporter plasmid pTA3-luc or pT81-luc and the pRL-CH110 control plasmid as described in the text. After 2 days, the cells were harvested and assayed for firefly luciferase activity with the dual luciferase system (Promega). The luciferase activities were normalized to Renilla luciferase activity, and the activation was obtained by setting the value of empty vector at 1.0. Shown are the means of activation transcriptional potential from three independent experiments.