Abstract
The response regulator CtrA controls chromosome replication by binding to five sites, a, b, c, d, and e, inside the Caulobacter crescentus replication origin (Cori). In this study, we demonstrate that integration host factor (IHF) binds Cori over the central CtrA binding site c. Surprisingly, IHF and CtrA share DNA recognition sequences. Rather than promoting cooperative binding, IHF binding hinders CtrA binding to site c and nearby site d. Unlike other CtrA binding sites, DNA mutations in the CtrA c/IHF site uniquely impair autonomous Cori plasmid replication. These mutations also alter transcription from distant promoters more than 100 bp away. When the CtrA c/IHF site was deleted from the chromosome, these cells grew slowly and became selectively intolerant to a CtrA phosphor-mimic allele (D51E). Since CtrA protein concentration decreases during the cell cycle as IHF protein concentration increases, we propose a model in which IHF displaces CtrA in order to bend Cori and promote efficient chromosome replication.
The dimorphic (swarmer and stalked) cell cycle of Caulobacter crescentus provides a valuable model to study chromosome replication control (32). Only the stalked cells perform chromosome replication. The swarmer cells must differentiate into stalked cells prior to chromosome replication (Fig. 1). A response regulator protein, cell cycle transcription regulator A (CtrA), orchestrates many cell cycle events (40). As illustrated in Fig. 1, CtrA accumulates in the swarmer cells and in the dividing cells, but CtrA is degraded during cell differentiation and therefore CtrA is absent in the stalked cells (11). CtrA is also phosphorylated in the swarmer cells and in the dividing cells (11, 23). These cell cycle patterns of CtrA protein phosphorylation, protein synthesis, and protein degradation regulate the transcription of ≈25% of the cell cycle genes (27).
FIG. 1.
Dimorphic cell cycle of C. crescentus. The cell cycle (28, 32, 39) and cell type distributions of both the CtrA (11) and the IHF (15) proteins are illustrated. Nonreplicating and replicating chromosomes are shown as figure-eight and theta-form shapes inside the swarmer and stalked cells, respectively. CtrA protein (indicated by gray shading) is abundant in the swarmer cells, degraded in the stalked cells, and resynthesized in the predivisional cells. IHF protein is not detected in swarmer cells but increases in stalked cells and predivisional cells.
CtrA also regulates chromosome replication by binding to five sites, designated a, b, c, d, and e, with the TTAA-N7-TTAA consensus motif (41, 45) inside the C. crescentus replication origin (Cori) (Fig. 2A). CtrA binding sites a and b overlaps the Cori AT-rich region and an exceptional “strong promoter” (Ps) whose transcription coincides with chromosome replication in the stalked cells (30). Previous studies suggested that CtrA binding at sites a and b represses Ps transcription, which in turn blocks Cori plasmid replication (30). CtrA binding at site d also represses Cori plasmid replication (41), but the mechanism of repression through site d is not known. CtrA binding site e is adjacent to an essential DnaA box (31), and site e is also near a promoter (P3) that transcribes the RP001 homologue (2). CtrA binding at e may influence replication by altering DnaA binding (19) and/or P3 transcription (32). CtrA binding site c is in the middle of the autonomous replicating Cori region (Fig. 2A), but its significance has not been investigated. In this study, we demonstrate that CtrA binding site c overlaps an integration host factor (IHF) binding site.
FIG. 2.
(A) C. crescentus chromosome replication origin (Cori). The thick horizontal line represents the minimal DNA sequences required for autonomous plasmid replication. Ps and P3 represent the two promoters whose transcription was assayed in Fig. 7. Open boxes labeled hemE and RP001 mark open reading frames. The horizontal bars mark the five CtrA binding sites a, b, c, d, and e, and their relative spacing is indicated by the corresponding nucleotide numbers for overlapping restriction sites for BglII (Bg), EcoRI (E), and HpaI (H). The solid arrow designates the essential DnaA box (based on Quon et al. [40]). The IHF binding site is illustrated by the triangle. (B) Precise IHF and CtrA footprints. Expanded view of Cori and DNase I footprint summary showing the exact nucleotides protected by separate binding of IHF (boxed nucleotides) or CtrA (double underlined nucleotides). To determine the exact nucleotides protected on the top strand, pGM1654 was 32P end labeled at the BamHI linker placed close to the IHF site and cleaved with XhoI to yield a ≈350-bp fragment with sites c, d, and e. Likewise, to determine the exact nucleotides protected on the bottom strand, pGM1654 was 32P end labeled at the EcoRI site and cleaved with PstI (+1) to yield a ≈450-bp fragment with sites a, b, and c. Otherwise, the footprint reaction conditions were those provided in Fig. 3 and 4. The bp +386 through +419 that were deleted in the Δ-Cori c/IHF strain, studied in Fig. 8 and 9, are also indicated. (C) The proposed IHF binding site. The 10 most conserved nucleotides within E. coli IHF binding sites (18) are shown aligned with Cori. The four most conserved nucleotides are in uppercase. The nucleotides that match Cori DNA are underlined and presumably form the binding site.
Escherichia coli IHF is a small heterodimeric “histone-like” protein that assists DNA recombination (8), transposition (6), transcription (7), and chromosome replication (25). IHF assists these diverse processes by binding to a specific DNA site and bending the DNA approximately 180 degrees (43). In a purified protein system, E. coli IHF assists DnaA, the major replication initiator protein, to unwind the AT-rich region of oriC, the chromosome replication origin (22). Interestingly, IHF binds E. coli oriC selectively at the start of chromosome replication in vivo (4, 5). IHF also redistributes oriC-bound DnaA protein, and this is apparently required to trigger chromosome replication (20, 44). However, despite these important studies of IHF in the E. coli oriC, the role of IHF in other chromosome replication origins, including C. crescentus Cori, has not been investigated.
C. crescentus IHF, like CtrA, appears to be a global cell cycle regulator. For example, the C. crescentus IHF (himA) and dnaA genes are transcribed in swarmer cells in apparent anticipation of chromosome replication (27). As illustrated in Fig. 1, IHF protein is undetectable in swarmer cells and rises in the stalked and predivisional cells (15, 16). Since CtrA protein is degraded when swarmer cells change into stalked cells (11), IHF protein increases when CtrA protein decreases (Fig. 1). The C. crescentus and E. coli IHF proteins are very homologous (36), and they have virtually identical biochemical properties. The C. crescentus IHF protein replaced the E. coli IHF in an in vitro lambda recombination assay (15). This assay is very demanding because it requires precise DNA sequence recognition as well as precise DNA bending. Therefore, the E. coli and C. crescentus IHF proteins are functionally interchangeable. IHF is required for cell cycle transcription of C. crescentus flagellum promoters (15, 16), and we speculated that IHF might also serve specific cell cycle functions during chromosome replication.
In this study we demonstrate that IHF binds the C. crescentus replication origin (Cori) over the central CtrA binding site c (Fig. 2). Surprisingly, IHF and CtrA have overlapping recognition sequences, and IHF binding hinders CtrA binding to site c. DNA mutations in the CtrA c/IHF site impair autonomous Cori plasmid replication, and they also alter transcription from distant promoters Ps and P3. To further test its physiological significance, the CtrA c/IHF site was deleted from the chromosome. These cells grew slowly and became uniquely intolerant to a CtrA phosphor-mimic allele (D51E). Since CtrA protein abundance decreases as IHF protein increases, we propose that IHF displaces CtrA inside Cori at the start of S phase and that this step promotes efficient chromosome replication.
MATERIALS AND METHODS
Bacterial strains, cultures, and analysis.
The strains and plasmids used in this study are listed in Table 1. C. crescentus cells were maintained on PYE medium (12) with glucose (0.02%). For in vivo transcription studies, plasmids were mobilized to C. crescentus from E. coli S17-1 (46) by conjugation (38). C. crescentus cells were grown exponentially in M2G medium, and β-glucuronidase and β-galactosidase assays were performed as described previously (38). For in vivo replication (autonomous replication) studies, plasmids were introduced into C. crescentus by electroporation (14).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | References or source |
|---|---|---|
| E. coli | ||
| BL21 | F−lon ompT hsdSB | Studier and Moffatt (46a) |
| S17-1 | E. coli 294::RP4-2(Tc::Mu)(Km::Tn7) | Simon et al. (46) |
| C. crescentus | ||
| NA1000 | Formerly CB15N, synchronizable wild type | Evinger and Agabian (12a) |
| NA1000 Δbla | Ampicillin-sensitive NA1000 | Marczynski and Shapiro (31) |
| Δ-Cori c/IHF | NA1000 Cori deletion +386/+419/BamHI | This work |
| Plasmidsa | ||
| pTrc7.4 | His6 N-terminally tagged CtrA protein | Quon et al. (40) |
| pKJH5 | MBP-EnvZ kinase protein fusion | Huang et al. (21) |
| pGM1270 | Cori BamHI fragment in pUC19 | Quon et al. (41) |
| pGM1413 | Cori deletion +386/+419/BamHI linker | Marczynski et al. (30) |
| pGM1654 | Cori deletion +240/+320/BamHI linker | Marczynski et al. (30) |
| pGM1877 | Cori HindIII at +214 to XhoI in pSK(+) | Siam and Marczynski (45) |
| pGM2512 | c-394 Cori BamHI in pUC19 | This work |
| pGM2513 | c-405 Cori BamHI in pUC19 | This work |
| pGM2514 | c-403 Cori BamHI in pUC19 | This work |
| pRSZ6-KanSacB | IncP oriT gusA lacZ reporter | Ouimet and Marczynski (37) |
All plasmids encode ampicillin resistance except pRSZ6-KanSacB, which encodes tetracycline resistance.
For CtrA allele induction studies, C. crescentus cells were grown to saturation in M2G medium (12) at 30°C and then adjusted to an optical density at 660 nm of 0.1 by dilution into fresh medium. For transfer into M2X (0.2% xylose) medium, cells were washed twice in the same medium before dilution. CtrA protein concentrations were measured by immunoblotting with anti-CtrA antiserum (1:1,000) as described previously (11). The Δ-Cori c/IHF deletion was mobilized from plasmid pGM1413 onto the chromosome of NA1000 by two-step homologous recombination, as described previously (29). The Δ-Cori c/IHF deletion is tagged with the BamHI linker GGGGATCCCC, and its placement inside Cori was monitored by Southern blotting (29).
Protein purification and phosphorylation.
Histidine-tagged CtrA fusion protein was overexpressed in E. coli BL21 (Novagen) from pTRC7.4 and purified as described previously (40). Maltose-binding protein (MBP)-EnvZ was purified from E. coli BL21 containing plasmid pKJH5 as described previously (21). CtrA was phosphorylated with purified MBP-EnvZ as described previously (45). Pure E. coli IHF protein was supplied by Steven Goodman and prepared by the protocol of Nash et al. (35).
DNase I footprint assays, site-directed DNA mutagenesis, and Southern blotting.
DNase I protection assays (13) were performed as described previously (45). The DNA fragments employed were end labeled with [γ-32P]dATP and cut with restriction enzymes, as specified in the figure legends, to yield short fragments that were isolated by gel electrophoresis (45). Precise IHF and CtrA footprints for Fig. 2B were obtained as follows. To determine the exact nucleotides protected on the top strand, pGM1654 was 32P end labeled at the BamHI linker located ≈50 bp to the left of the IHF site and cleaved with XhoI to yield a ≈350-bp fragment with sites c, d, and e. Likewise, to determine the exact nucleotides protected on the bottom strand, pGM1654 was 32P end labeled at the EcoRI site and cleaved with PstI (+1) to yield a ≈450-bp fragment with sites a, b, and c. Otherwise, the footprint reaction conditions were those provided in Fig. 3 and 4.
FIG. 3.
(A) CtrA and IHF binding in Cori. DNase I footprint assays used 0.02 μM CtrA≈P or 0.05 μM IHF (+ lanes). Purified CtrA was phosphorylated by MBP-EnvZ kinase, as described by Reisenauer et al. (42). Plasmid pGM1270 was 32P end labeled at the BglII site and cleaved with XhoI, yielding a fragment with CtrA binding sites b, c, d, and e. CtrA≈P and IHF binding to sites c and d is displayed. (B) IHF binding to mutant Cori DNA. DNase I footprint assays done as for A but employing mutant 32P-labeled Cori DNA fragments derived from plasmids pGM2512, pGM2513, and pGM2512 (Table 1). Their exact changes are shown in Fig. 5.
FIG. 4.
CtrA binding to Cori in the presence of IHF. DNase I footprint assays used 0.01 μM CtrA≈P (+ lanes). Plasmid pGM1877 was 32P end labeled at the artificial HindIII site and cleaved with XhoI, yielding a fragment with CtrA binding sites a, b, c, d, and e (as illustrated). Increasing concentrations of IHF protein (0.01 to 0.2 μM) were added simultaneously with 0.01 μM CtrA≈P. The horizontal arrowheads mark the flanking DNase I-hypersensitive sites that characterize CtrA footprints.
To determine Kd values for IHF and CtrA, equilibrium binding assays were performed with our standard footprinting protocol, as described previously (45). Under these conditions, where the molar concentration of protein greatly exceeds the molar concentration of target DNA, the Kd equals the protein concentration that provides 50% target site occupancy (13). The range of protein concentrations that span the 50% occupancy concentration was determined empirically from multiple footprint experiments analyzed by phosphorimaging and IQuant software (Molecular Dynamics).
Site-directed DNA mutagenesis of Cori CtrA binding site c was performed with the U.S.E. mutagenesis kit (Pharmacia), which employs the “unique site elimination” mutagenesis protocol. We mutagenized double-stranded Cori plasmid pGM1270 with these oligonucleotides: Mut c-394, CTTCCCACATGGGGATCCCGCTCTGTTAATC; Mut c-405, GGTTAACGCTCTGGAATTCATGGGGATGAATG; and Mut c-403, CATGGGGTTAACGCTCACTTAATCATGGGGATG. All mutations were confirmed by Sanger dideoxy sequencing with the T7 sequencing kit (Pharmacia).
Southern blot analysis was also performed to monitor plasmid replication. C. crescentus strain NA1000 Δbla was electroporated (14) with pUC19-based Cori plasmids (Table 1) and plated on PYE plus ampicillin (20 μg/ml). Cells were harvested by washing these plates with PYE medium. The cell density was adjusted to an optical density of 5.0 by dilution, and total DNA was prepared, digested with BamHI endonuclease, and analyzed by Southern blotting, as described previously (30). The BamHI Cori DNA fragment from plasmid pGM1270 was used as the hybridization probe (30).
RESULTS
IHF binds the C. crescentus replication origin (Cori).
Although CtrA binds cooperatively to adjacent sites a and b, CtrA binds independently to distant sites c, d, and e (45). IHF-induced DNA bending often promotes cooperative protein binding because it allows distal bound proteins to contact each other (17, 43). We therefore tested whether IHF might promote additional cooperative CtrA binding. E. coli IHF protein is interchangeable with C. crescentus IHF protein (15) and was therefore used in our DNase I footprint assays. Accordingly, we first tested IHF binding to 32P-end-labeled DNA that spans all five Cori CtrA binding sites. IHF selectively bound Cori at one site with a Kd of ≈20 nM. As illustrated in Fig. 2A, this IHF site overlapped CtrA binding site c (Fig. 3A and 4 and data not shown).
CtrA site c/IHF footprints have overlapping recognition sequences.
We determined the precise footprint boundaries for both CtrA and IHF. To obtain single-nucleotide resolution, Cori fragments were 32P end labeled close to site c/IHF at position +332 (top strand) or at position +452 (bottom strand). Otherwise, the same footprint protocol was repeated as in Fig. 3A (data not shown), and the Fig. 2B diagram summarizes our results. The exact nucleotides protected from DNase I digestion by IHF binding (boxed nucleotides) and likewise protected by CtrA binding (double underlined nucleotides) are shown in Fig. 2B. The IHF footprint spans the CtrA footprint at site c and extends on both sides. The CtrA consensus sequence (TTAA-N7-TTAA) is centered within the CtrA footprint. Likewise, the proposed IHF binding sequence (underlined in Fig. 2C) is centered within the IHF footprint. A comparison between this CtrA consensus sequence and this proposed IHF binding sequence (18) implies that the CtrA and IHF recognition sequences overlap by at least 4 critical bp. Therefore, these results (Fig. 2BC) suggest that CtrA and IHF cannot occupy this site at the same time.
Antagonistic binding of IHF and CtrA inside Cori.
We tested how IHF influences CtrA binding to site c and to the distal CtrA binding sites. Our initial hypothesis was that IHF might aid CtrA binding. We therefore combined various concentrations of phosphorylated CtrA (CtrA≈P) and IHF in our DNase I footprint reactions with 32P end-labeled Cori fragments that contain CtrA sites b, c, d, and e, as in Fig. 3, and all five CtrA sites a to e, as in Fig. 4. In these and other footprint experiments, subsaturating concentrations of CtrA and CtrA≈P were used so that increased CtrA binding could be detected by adding IHF protein. However, IHF did not increase the binding of CtrA or CtrA≈P to Cori. Instead, IHF always diminished CtrA binding to site c, apparently by direct displacement, as suggested by our previous sequence analysis (Fig. 2BC). For example, diminished CtrA binding to site c occurs when increasing amounts of IHF are added to CtrA≈P (Fig. 4 and data not shown). At high IHF concentrations, the footprint pattern at site c in the IHF plus CtrA≈P lanes resembles the IHF-only footprint. CtrA≈P binding at site d was also diminished in the CtrA≈P plus IHF lanes compared to the CtrA≈P-only lanes (Fig. 4). Apparently, IHF binding at site c also hinders CtrA≈P binding at site d. Another indication of weaker CtrA binding is the absence of CtrA protein-induced DNase I-hypersensitive sites flanking CtrA binding sites c and d (Fig. 4).
Directed mutations selectively reduce binding of CtrA and IHF to Cori site c.
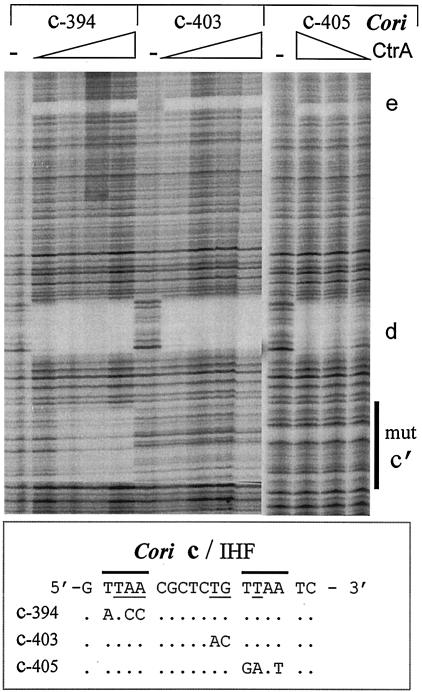
To further investigate CtrA and IHF binding at site c, we created three mutants (c-394, c-403, and c-405; Fig. 5) and determined their binding affinities for CtrA, CtrA≈P, and IHF. The Cori nucleotides that match the CtrA and the IHF consensus binding sites are shown aligned with c-394, c-403, and c-405 in Fig. 5. Although the CtrA and IHF binding sites overlap, this sequence analysis suggested that mutation c-403 might reduce IHF binding without reducing CtrA binding. Accordingly, DNase I footprint assays were performed with Cori fragments containing the wild-type DNA and each of these three mutations. Mutation c-394, which changes 3 bp in the left TTAA motif of CtrA binding site c, only bound CtrA≈P with a Kd of 50 nM. Therefore fivefold more CtrA≈P is required to occupy c-394 than wild-type site c (Fig. 5). Likewise, fivefold more unphosphorylated CtrA is required to occupy c-394 (Kd = 200 nM) than wild-type site c (data not shown). However, we did not observe any binding of CtrA or CtrA≈P to c-405, which changed 3 bp in the right TTAA motif of site c, or to c-403, which changed 2 bp between the TTAA motifs of c. The Kd for CtrA≈P binding to c-403 and c-405 must exceed 2 μM. As expected, these three mutations did not affect CtrA and CtrA≈P binding to sites d and e, and these sites served as internal controls for quantitative CtrA binding to site c. Also, strong and unaltered CtrA≈P binding to sites d and e on all three Cori mutants (Fig. 5) supports our earlier conclusion that CtrA binding to sites c, d, and e is independent.
FIG. 5.
DNase I footprint assays of the three Cori site c/IHF mutations. DNase I footprint assays done as for Fig. 3A but employing mutant 32P-labeled Cori DNA fragments derived from plasmids pGM2512 (c-394), pGM2513 (c-405), and pGM2514 (c-403) (Table 1). Their exact changes are shown below and are aligned with the wild-type Cori DNA sequences. The overlined nucleotides match the CtrA consensus, and the underlined nucleotides match the IHF consensus. Increasing concentrations of CtrA≈P (0.02 to 0.5 μM) were added to each footprint reaction. The autoradiograms display the Cori DNA spanning CtrA binding sites c, d, and e. The bar marked mut c′ designates the mutated sequences presented below.
IHF binding to site c mutations c-394, c-403, and c-405 (Fig. 5) was weaker than to the wild-type Cori by approximately fivefold (Fig. 3B and data not shown). Due to the low binding affinities and protein aggregation at high IHF concentrations, we were unable to derive accurate Kd values from our DNase I footprint assays. However, qualitatively site c mutations c-405, c-403, and c-394 created progressively weaker IHF binding sites.
In vivo, Cori plasmid replication correlates with in vitro CtrA binding to site c.
We tested how altered IHF and CtrA binding at site c might affect replication. Plasmids bearing wild-type Cori and Cori with mutation c-394, c-403, or c-405 (Fig. 5) were introduced into C. crescentus cells by electroporation. Plating on PYE plus ampicillin selects for cells that support autonomous Cori plasmid replication (30). The absence of CtrA binding and weak IHF binding to c-405 and c-403 correlates with no replication, as evidenced by no detectable colonies in this assay. Partial CtrA and weak IHF binding to c-394 correlates with fewer colonies as well as smaller colonies than the wild-type Cori plasmid control. This result was supported by Southern blot analysis of DNA extracted from these transformed cells. Compared to wild-type Cori, weaker CtrA and IHF binding to Cori c-394 gave ≈2-fold fewer autonomously replicating Cori plasmids per chromosome (Fig. 6).
FIG. 6.
Autonomous replication assays. Mutant Cori plasmids pGM2512 (c-394), pGM2513 (c-405), and pGM2514 (c-403) and wild-type Cori plasmid pGM1270 (Table 1) were electroporated into ampicillin-sensitive C. crescentus NA1000 (Δbla) cells and spread over PYE (20 μg of ampicillin per ml) plates. Autonomous replication was assayed by CFU (we obtained 105 CFU per ng of Cori plasmid), and these results are summarized by + for wild-type Cori, by +/− for fewer than 50% CFU as well as smaller colonies compared to wild-type Cori, and by − for the absence of colonies. Total cellular DNA was extracted from these colonies for analysis by Southern blotting and hybridization with 32P-labeled Cori DNA, as described in Materials and Methods. The marked bands correspond to the linear Cori plasmids and to the 1.6-kb chromosomal Cori fragment.
Mutations in the c/IHF site selectively affect distant transcription promoters.
Since IHF binding creates a “U-shaped” DNA bend (43), the central position of the c/IHF site inside Cori (Fig. 2) suggests that both ends might juxtapose and influence distant DNA and protein interactions. Transcription from the Ps and P3 promoters is directed outwards at both ends of Cori, and it is easily assayed by fusion to flanking transcription reporter genes, as illustrated in Fig. 7. We therefore tested how altered IHF and CtrA binding at site c might affect Ps and P3 transcription. Autonomously replicating (IncP replicon) transcription reporter plasmids bearing wild-type Cori DNA and the same span of Cori DNA with mutations c-394, c-403, and c-405 (Fig. 5) were introduced into C. crescentus cells. The corresponding enzyme activities, β-glucuronidase (Ps::gusA) and β-galactosidase (P3::lacZ), were measured in exponentially growing cultures and are displayed as bar graphs (Fig. 7). Compared to efficient wild-type Cori transcription, mutation c-403 moderately decreased distant P3 transcription (Fig. 7). However, mutations c-394 and c-405 reciprocally increased Ps transcription ≈2-fold and decreased P3 transcription ≈4-fold (Fig. 7). Therefore, these central mutations selectively influence distant transcription from both ends of Cori.
FIG. 7.
In vivo transcription from Cori Ps and P3 promoters. Cori wild-type DNA and Cori DNA with mutations c-394, c-403, and c-405 (Fig. 5) were mobilized onto transcription reporter plasmids by homologous recombination in E. coli (38). As illustrated, these manipulations fused Cori DNA sequences spanning positions +214 and +998 (Fig. 2) between the gusA and lacZ reporter genes of IncP plasmid pRSZ6-KanSacB. These plasmids were then mobilized into C. crescentus strain NA1000. The corresponding enzyme activities for the Ps::gusA and P3::lacZ strains were measured in exponentially growing M2G cultures and are displayed as bar graphs. Hatched bars present β-glucuronidase activity, and solid bars present β-galactosidase activity (in Miller units).
Deleting the Cori c/IHF site impairs cell growth.
To test the physiological relevance of the Cori site c/IHF site, we deleted the DNA between positions +386 and +419 (shown in Fig. 2B) and introduced this construct (Δ-Cori c/IHF in Fig. 8A) into wild-type C. crescentus by homologous DNA recombination. As described in Materials and Methods, Δ-Cori c/IHF is tagged with the BamHI linker DNA. This tag allowed a Southern blot screen for strains that replaced the wild-type Cori with Δ-Cori c/IHF (Fig. 8A) by double (flanking) homologous DNA recombination.
FIG. 8.
(A) DNA structure of mutant Cori on the chromosome of strain Δ-Cori c/IHF. This strain was derived from wild-type (Wt) C. crescentus NA1000 by homologous recombination, as described in Materials and Methods. (B) Growth curves for NA1000 (wild type) and Δ-Cori c/IHF strains. Freshly saturated cultures were diluted into M2G (minimal glucose) medium, and their optical density (O.D.) was measured every 2 h.
The Cori c/IHF site is clearly not essential for cell viability. Following selection for double homologous recombination, we recovered wild-type Cori and Δ-Cori c/IHF strains with equal frequency, and these cells were indistinguishable by 1,000× light microscopy, indicating that the general program of dimorphic cell division was not disturbed (data not shown). However, the Cori c/IHF site is required for efficient cell growth, since a Δ-Cori c/IHF strain grows ≈4-fold slower than the wild type in standard minimal glucose (M2G) medium (Fig. 8B).
Deleting the c/IHF site causes intolerance to a CtrA phosphor-mimic allele.
Since CtrA is an essential cell cycle regulator that directly binds and represses Cori, we also tested how mutant Cori cells (Δ-Cori c/IHF in Fig. 8) respond to increased concentrations of CtrA protein and to different CtrA alleles. While the wild-type CtrA protein is degraded when the swarmer cells change into stalked cells (Fig. 1), the CtrAΔ3 allele (encoding a 3-amino-acid terminal deletion) is not degraded, and it remains abundant in all cell types (11). Nonetheless, an oversupply of CtrAΔ3 permits a normal cell cycle, including stalked cell chromosome replication, because cell cycle protein phosphorylation regulates CtrAΔ3 activity (11). In contrast, the CtrAD51E allele (an aspartate to glutamate change at the phosphorylation site) is not phosphorylated, but CtrAD51E is termed a phosphor-mimic because it resembles the phosphorylated form of CtrA (11). An oversupply of CtrAD51E also permits a normal cell cycle, because cell cycle protein degradation regulates CtrAD51E activity (11). Blocking chromosome replication requires the double mutant CtrAΔ3D51E. An oversupply of CtrAΔ3D51E blocks chromosome replication, because CtrAΔ3D51E is an active phosphor-mimic that cannot be removed (11).
We reproduced these reported results for the wild-type Cori cells, and we demonstrated an unexpected sensitivity to the CtrAD51E allele for the Δ-Cori c/IHF strain (Fig. 8). As reported previously (11), we also employed the same plasmids bearing different CtrA alleles whose synthesis is controlled by the xylose-inducible promoter (Fig. 9A). These plasmids were first introduced into wild-type C. crescentus (wild-type Cori) cells in the presence of glucose to repress transcription from the xylose promoter (33). As a control, we measured CtrA protein synthesis and accumulation after 8 h of growth in glucose and in xylose medium (Fig. 9A). This immune blot demonstrated that xylose induced the synthesis of these CtrA proteins, and it also confirmed that high concentrations of CtrAΔ3 and CtrAD51E are tolerated. As previously reported (11), xylose induction did not block the cell cycle in these wild-type Cori cells except for the CtrAΔ3D51E double mutant allele (Fig. 9B). The partial cell cycle block upon xylose induction of CtrAΔ3 (Fig. 9B) was also observed previously (11). A high concentration of CtrAΔ3 (Fig. 9A) causes this relatively weak cell division defect. These cells are longer and divide more slowly but otherwise show wild-type behavior (11).
FIG. 9.
(A) Immune blot of CtrA allele-induced cultures. As described previously in the text, cultures in M2G (G, 0.2% glucose) or M2X (X, 0.2% xylose) were grown exponentially for 8 h before analysis for CtrA protein concentration by immune blotting with anti-CtrA serum, as described in Materials and Methods. (B) Summary of growth and microscopic (cell morphology) assays. +, wild-type growth and appearance; +/−, reduced growth and elongated cells; −, no growth or blocked cell division following a shift from M2G to M2X medium. (C) Dark-field microscopy (1,000×) illustrating blocked cell division following a shift from M2G to M2X medium. The Δ-Cori c/IHF xylose-inducible CtrA D51E strain was grown in M2G, and then half of this culture was shifted to M2X for 4 h prior to photography.
Enhanced sensitivity to the CtrA51E allele was first demonstrated when we repeated these experiments with the Δ-Cori c/IHF strain (Fig. 8) and found that we could not establish the CtrAΔ3D51E double mutant plasmid in the presence of either glucose or xylose (Fig. 9B). Since this plasmid is easily established in wild-type Cori cells when glucose represses transcription (Fig. 9B), this negative result implies an exceptional intolerance for CtrAΔ3D51E by the Δ-Cori c/IHF strain. In contrast, the wild-type CtrA-, CtrAΔ3-, and CtrAD51E-expressing plasmids were easily established in the Δ-Cori c/IHF strain in the presence of glucose. When these cells were shifted to xylose medium, inducing CtrAD51E selectively blocked the cell cycle (Fig. 9B). The morphology of the Δ-Cori c/IHF strain containing the CtrAD51E plasmid in glucose culture resembles wild-type cells (Fig. 9C). However, when this culture was shifted to xylose medium, after 4 h these cells stopped dividing and presented a filamentous morphology (Fig. 9C), typical of cells with blocked chromosome replication (19). Pulse-labeling with 32P confirmed that these cells reduced DNA synthesis (data not shown). This cell cycle block is specific to the CtrAD51E allele, since CtrAΔ3 is induced to greater concentrations (Fig. 9A) without causing a comparable block (Fig. 9B).
DISCUSSION
Overlapping IHF and CtrA binding inside the C. crescentus replication origin.
We demonstrated that IHF binds the C. crescentus replication origin (Cori) over the central CtrA binding site c (Fig. 2). Surprisingly, IHF and CtrA have overlapping recognition sequences. This conclusion is supported by the overlapping DNase I protection patterns presented in Fig. 2B and Fig. 3A. Centered within these footprint boundaries, the consensus sequence (37) for CtrA (TTAA-N7-TTAA) and the consensus sequence (18) for IHF (TAA-T-TTGATT) share 4 bp (Fig. 2C). Although this consensus sequence for IHF matches the Cori DNA in 6 of 10 positions (italic), as noted in Fig. 2C, Cori contains all 4 bp most frequently found at IHF binding sites (18). These consensus sequences clearly specify protein binding, because site-directed base pair changes (Fig. 5) reduce or abolish binding by IHF (Fig. 3B) and by CtrA (Fig. 5). However, additional sequences must specify CtrA binding. For example, c-403 was originally designed to block IHF binding without blocking CtrA binding, since the c-403 changes the most conserved IHF consensus base pairs that lie outside the CtrA consensus (Fig. 5). However, c-403 blocks both IHF (Fig. 3B) and CtrA protein binding (Fig. 5). Therefore, c-403 identifies addition recognition sequences used by CtrA. Previous studies also argued that the intervening 7 bp influenced CtrA binding (37), but the complete absence of CtrA binding to c-403 was unexpected. Considering these overlapping binding requirements, we subsequently combined and designated only one CtrA Cori c/IHF binding site.
Cooperative CtrA binding inside the C. crescentus replication origin.
Our initial hypothesis that IHF might promote cooperative CtrA binding was not supported by our in vitro binding experiments. Instead, as suggested by their overlapping binding sites (Fig. 2), IHF binding hinders CtrA binding to site c. Hindrance without increased CtrA affinity for Cori DNA was observed in Fig. 4 and in similar experiments (data not shown) where IHF protein was added to CtrA protein at subsaturating concentrations with respect to all five Cori CtrA binding sites. Nonetheless, our CtrA protein preparations were certainly capable of cooperative binding over short distances. We previously demonstrated that CtrA protein exhibits two modes of cooperative binding inside Cori and that both modes are enhanced by CtrA phosphorylation (45). CtrA proteins cooperate to increase affinity across two TTAA “half-sites” with N7 spacing (42, 45). Likewise, CtrA proteins cooperate across two complete TTAA-N7-TTAA Cori sites, a and b, with N6 spacing (45). However, cooperative binding over longer distances across sites a to e spanning Cori was not observed with either CtrA≈P (Fig. 4) or unphosphorylated CtrA protein (data not shown). If long-distance cooperative binding occurs inside Cori, we presume that it cannot be reconstituted in vitro with purified IHF and CtrA proteins.
Despite negative in vitro binding results, long distance cooperative CtrA binding is suggested by our in vivo transcription experiments (Fig. 7). Self-replicating broad-host-range (IncP) plasmids maintain both wild-type and mutant (nonreplicating) Cori DNA in C. crescentus, and this allowed us to assay Cori transcription separately from replication. These plasmids carry transcription reporter genes (38) that monitor the Ps and P3 promoters at both ends of Cori. We were surprised that mutations at site c/IHF, in the middle of Cori, could simultaneously increase Ps transcription and decrease P3 transcription (Fig. 7). Interestingly, Ps transcription is repressed by CtrA (40, 41), and P3 transcription is stimulated by CtrA (unpublished results). Perhaps mutations that weaken CtrA binding at site c (Fig. 5) likewise weaken CtrA binding at distant CtrA binding sites. Ps overlaps CtrA sites a and b (30). Since Ps transcription starts at position +251 (Fig. 2), it is hard to explain how mutations at +394, +403, and +405 (Fig. 5) influence Ps transcription without DNA bending or looping. However, it is not clear if the DNA bending or looping involves CtrA and IHF. Our inability to reconstitute cooperative binding with CtrA and IHF suggests that this phenomenon is more complicated and probably involves additional replication proteins.
Unique impairment of replication by mutations in the CtrA c/IHF site.
DNA mutations in the CtrA c/IHF site impair autonomous Cori plasmid replication (Fig. 6). These results suggest that Cori plasmid replication requires CtrA binding at site c. Comparing Fig. 5 and 6, partial CtrA binding to mutation c-394 correlates with partial replication, and the lack of CtrA binding to mutations c-403 and c-405 correlates with the lack of replication. These mutations also reduce IHF binding to the CtrA c/IHF site (Fig. 3B), so reduced IHF binding also correlates with reduced or absent replication. However, this correlation is not exact, since the lack of IHF binding to c-394 associates with partial replication, but partial IHF binding to c-403 and c-405 associates with absolutely no replication.
It was unexpected that Cori plasmid replication should require CtrA binding at site c. CtrA is a proposed repressor of chromosome replication in swarmer cells (41). According to this simple model, mutations that only block CtrA binding should increase Cori plasmid replication. As predicted, mutations at site d reduce CtrA binding and increase Cori plasmid replication (41). Similarly, mutations at sites a, b, and e can reduce or abolish CtrA binding and still maintain autonomous Cori plasmid replication (30). Perhaps the unique combination of CtrA and IHF binding to this central Cori DNA accounts for the severity of the c-394, c-403, and c-405 mutations.
Physiological significance of CtrA c/IHF binding site.
To further explore how C. crescentus uses the CtrA c/IHF site, we deleted the DNA that spans both CtrA and IHF footprints, between +386 and +419 (Fig. 2B), from the chromosome (Fig. 8). Although these strains were viable, they grew slowly (Fig. 8B), and they became uniquely intolerant to CtrAD51E (Fig. 9BC). Preliminary experiments also suggest that the swarmer to stalked-cell phase remains the same, because we did not observe extra swarmer cells during logarithmic growth. However, the cells take longer to divide, and we speculate that this is due to inefficient coordination between cell division and chromosome replication.
We were first surprised that this deletion permits replication when placed by homologous recombination into the whole-chromosome context, because it abolishes autonomous replication in the Cori plasmid context (data not shown). Note that changes c-403 and c-405 also abolish autonomous replication (Fig. 6). Apparently, Cori plasmid replication is more sensitive to mutations, and the whole chromosome provides compensatory functions. Similar results are observed for E. coli oriC plasmids and its chromosome (47).
The CtrAD51E allele-specific intolerance is also surprising and very conspicuous. Even when substantially higher concentrations of CtrAΔ3 are induced by xylose (Fig. 9A), the Δ-Cori c/IHF strain shows only minor growth defects (e.g., elongated cells) that are indistinguishable from those induced in the wild-type strain (Fig. 9B). The CtrAΔ3 protein retains full function in vivo since low concentrations of CtrAΔ3 fully complement a ctrA null allele (41). In contrast, CtrA D51E is only partially functional in vivo. CtrA D51E cannot complement a ctrA null allele unless CtrA D51E is supplied at high concentrations (11), such as by the xylose-induced promoter (Fig. 9). CtrA D51E is not phosphorylated in vivo (11) or in vitro (42), and CtrA D51E has a low affinity for target DNA that is indistinguishable for wild-type unphosphorylated CtrA (unpublished results). Nonetheless, CtrA D51E is clearly a gain-of-function allele resembling the phosphorylated protein in vivo (11). For example, the CtrA D51E allele uniquely bypasses a requirement for its cognate kinase (23), but its acquired or altered biochemical properties remain to be determined.
Our results constrain the requirements for IHF in C. crescentus chromosome replication. CtrA and IHF have overlapping recognition sequences (Fig. 2, 3, and 5), and it is not possible to unambiguously separate the requirements for either protein. However, if IHF does bind Cori in vivo, then the Δ-Cori c/IHF strains clearly demonstrate that IHF binding is not absolutely essential. Slower growth and altered interactions with CtrA D51E suggest that IHF is an auxiliary protein that makes replication and replication controls more efficient. Likewise, in E. coli, IHF is not absolutely essential for chromosome replication (1). E. coli IHF serves auxiliary roles that are currently viewed as promoting favorable contacts between oriC DNA and other replication proteins (34).
Comparison with other bacterial replication models.
Most chromosome replication models are based on the E. coli in vitro replication system where replication from the cloned replication origin (oriC) is reconstituted from purified enzymes (9). In this system, E. coli DnaA protein, the major replication initiator protein, binds multiple DnaA boxes inside oriC, and E. coli IHF assists DnaA to unwind the AT-rich region of oriC. Subsequently, DnaA recruits the DnaB helicase to the unwound AT-rich region, and this is regarded as the commitment step for chromosome replication (34). Therefore, E. coli IHF assists DnaA immediately before the chromosome becomes committed to replication, but IHF is not an essential E. coli protein, and another histone-like protein (Hu) can replace IHF in vitro (10, 34). This is reasonable if the major role for IHF is to strategically bend DNA (17), and this might occur spontaneously or less efficiently through contacts with other proteins.
Many bacteria contain DnaA, and their replication origins contain multiple DnaA boxes that match the E. coli DnaA box consensus (24, 26). C. crescentus also requires DnaA for chromosome replication (19), but Cori lacks multiple conspicuous matches to the E. coli DnaA box consensus. The closest matching Cori sequence (8 of 9 bp) to the E. coli DnaA box consensus is shown in Fig. 2A. This sequence is required for autonomous replication (31), but it is on the opposite end of Cori and away from the Ps promoter and CtrA sites a and b that overlap the AT-rich region (31, 45). By analogy to E. coli, if this AT-rich region is also unwound by C. crescentus DnaA, then a strategic 180-degree bend from the middle of Cori might aid this contact. Interestingly, E. coli IHF binds oriC selectively at the start of chromosome replication in vivo (4). In C. crescentus, the programmed removal of CtrA combined with the synthesis of IHF in the stalked cells (Fig. 1) suggests that IHF might also selectively bind Cori during chromosome replication. This hypothesis is consistent with the antagonistic CtrA and IHF binding at site c in vitro (Fig. 4). As in E. coli, cell cycle C. crescentus protein binding can be tested by in vivo footprint techniques (4). Although Cori is the only established chromosome replication origin among the alpha-proteobacteria (32), an IHF binding site is also present in the center of the proposed Rickettsia prowazekii replication origin (3). Therefore, IHF may be widely used among bacteria as a strategic aid for chromosome replication.
Acknowledgments
We thank Steven Goodman for providing IHF protein.
This work was supported in part by Le Fonds pour la Formation de Chercheurs et l'Aide de Recherche (FCAR) as a Ph.D. scholarship to R.S. and A.K.C.B., by Medical Research Council of Canada (MRC/CIHR) grant MT-13453, and by MRC Scholarship Award SH-50791-AP007403 to G.T.M.
REFERENCES
- 1.Bates, D. B., T. Asai, Y. Cao, M. W. Chambers, G. W. Cadwell, E. Boye, and T. Kogoma. 1995. The DnaA Box R4 in the minimal oriC is dispensable for initiation of Escherichia coli chromosome replication. Nucleic Acids Res. 23:3119-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brassinga, A. K. C., R. Siam, and G. T. Marczynski. 2001. Conserved gene cluster at replication origins of the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii. J. Bacteriol. 183:1824-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brassinga, A. K. C., R. Siam, W. McSween, H. Winkler, D. Wood, and G. T. Marczynski. 2002. Conserved response regulator and IHF binding sites in the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosome replication origins. J. Bacteriol. 184:5789-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassler, M., J. Grimwade, and A. Leonard. 1995. Cell cycle-specific changes in nucleoprotein complexes at a chromosome replication origin. EMBO J. 14:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassler, M., J. Grimwade, K. C. McGarry, R. T. Mott, and A. Leonard. 1999. Drunken-cell footprints: nuclease treatment of ethanol-permeablilized bacteria reveals an initiation-like nucleoprtein complex in stationary phase replication origins. Nucleic Acids Res. 27:4570-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalmers, R., A. Guhathakurta, H. Benjamin, and N. Kleckner. 1998. IHF Modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93:897-908. [DOI] [PubMed] [Google Scholar]
- 7.Colland, F., M. Barth, R. Hengge-Aronis, and A. Kolb. 2000. Sigma factor selectivity of Escherichia coli RNA polymerase: role for Crp, IHF and Lrp transcription factors. EMBO J. 19:3028-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig, N. L., and H. A. Nash. 1984. E. coli integration host factor binds to specific sites in DNA. Cell 39:707-716. [DOI] [PubMed] [Google Scholar]
- 9.Crooke, E. 1995. DNA synthesis initiated at oriC: in vitro replication reactions. Methods Enzymol. 262:500-506. [DOI] [PubMed] [Google Scholar]
- 10.Dixon, N. E., and A. Kornberg. 1984. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc. Natl. Acad. Sci. USA 81:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1 to S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 12.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 12a.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galas, D. J., and A. Schmitz. 1978. DNase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 5:9357-9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilchrist, A., and J. Smit. 1991. Transformation of freshwater and marine caulobacters by electroporation. J. Bacteriol. 173:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by a 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gober, J. W., and L. Shapiro. 1990. Integration host factor is required for the activation of developmentally regulated genes in Caulobacter. Genes Dev. 4:1494-1505. [DOI] [PubMed] [Google Scholar]
- 17.Goodman, S. D., S. C. Nicholson, and H. A. Nash. 1992. Deformation of DNA during site-specific recombination of bacteriophage lambda: replacement of IHF protein by HU protein or sequence-directed bends. Proc. Natl. Acad. Sci. USA 89:11910-11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodrich, J. A., M. L. Schwartz, and W. R. McClure. 1990. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 18:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbatyuk, B., and G. T. Marczynski. 2001. Physiological consequences of blocked Caulobacter crescentus DnaA expression, an essential DNA replication gene. Mol. Microbiol. 40:485-497. [DOI] [PubMed] [Google Scholar]
- 20.Grimwade, J. E., V. T. Ryan, and A. C. Leonard. 2000. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 35:835-844. [DOI] [PubMed] [Google Scholar]
- 21.Huang, K.-J., C.-Y. Lan, and M. M. Igo. 1997. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc. Natl. Acad. Sci. USA 94:2828-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang, D. S., and A. Kornberg. 1992. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 267:23083-23086. [PubMed] [Google Scholar]
- 23.Jacobs, C., N. Ausmees, S. J. Cordwell, L. Shapiro, and M. T. Laub. 2003. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol. Microbiol. 47:1279-1290. [DOI] [PubMed] [Google Scholar]
- 24.Jakimowicz, D., J. Majka, G. Konopa, G. Wegrzyn, W. Messer, H. Schrempf, and J. Zakrzewska-Czerwinska. 2000. Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol. 298:351-364. [DOI] [PubMed] [Google Scholar]
- 25.Kano, Y., T. Ogawa, T. Ogura, S. Hiraga, T. Okazaki, and F. Imamoto. 1991. Participation of the histone-like protein HU and of IHF in minichromosome maintenance in Escherichia coli. Gene 103:25-30. [DOI] [PubMed] [Google Scholar]
- 26.Krause, M., and W. Messer. 1999. DnaA protein of Escherichia coli and Bacillus subtilis: coordinate actions with single-strand DNA-binding protein and interspecies inhibition during open complex formation at the replication origins. Gene 228:123-132. [DOI] [PubMed] [Google Scholar]
- 27.Laub, M. T., H. H. McAdams, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 28.Lott, T., N. Ohta, and A. Newton. 1987. Order of gene replication in Caulobacter crescentus: use of in vivo labeled genomic DNA as a probe. Mol. Gen. Genet. 210:543-550. [DOI] [PubMed] [Google Scholar]
- 29.Marczynski, G. T. 1999. Chromosome methylation and the measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J. Bacteriol. 181:1984-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marczynski, G. T., K. Lentine, and L. Shapiro. 1995. A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 9:1543-1557. [DOI] [PubMed] [Google Scholar]
- 31.Marczynski, G. T., and L. Shapiro. 1992. Cell cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J. Mol. Biol. 226:959-977. [DOI] [PubMed] [Google Scholar]
- 32.Marczynski, G. T., and L. Shapiro. 2002. Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Microbiol. 56:625-656. [DOI] [PubMed] [Google Scholar]
- 33.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messer, W., and C. Weigel. 1996. Initiation of chromosome replication, p. 1579-1601. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C.
- 35.Nash, H. A., C. A. Robertson, E. Flamm, R. A. Weisberg, and H. I. Miller. 1987. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J. Bacteriol. 169:4124-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouimet, M.-C., and G. T. Marczynski. 2000. Analysis of a cell cycle promoter bound by a response regulator. J. Mol. Biol. 302:761-775. [DOI] [PubMed] [Google Scholar]
- 38.Ouimet, M.-C., and G. T. Marczynski. 2000. Transcription reporters that shuttle between high-copy Escherichia coli plasmids and low-copy broad-host-range plasmids. Plasmid 44:152-162. [DOI] [PubMed] [Google Scholar]
- 39.Poindexter, J. S. 1981. The Caulobacters: Ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 41.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 95:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reisenauer, A., K. Quon, and L. Shapiro. 1999. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol. 181:2430-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice, P. A., S.-w. Yang, K. Mizuuchi, and H. A. Nash. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87:1295-1306. [DOI] [PubMed] [Google Scholar]
- 44.Ryan, V. T., J. E. Grimwade, C. J. Nievera, and A. C. Leonard. 2002. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 46:113-124. [DOI] [PubMed] [Google Scholar]
- 45.Siam, R., and G. T. Marczynski. 2000. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 19:1138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon, R., U. Priefer, and A. Puler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 46a.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 47.Weigel, C., W. Messer, S. Preiss, M. Welzeck, Morigen, and E. Boye. 2001. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 40:498-507. [DOI] [PubMed]