FIG. 2.
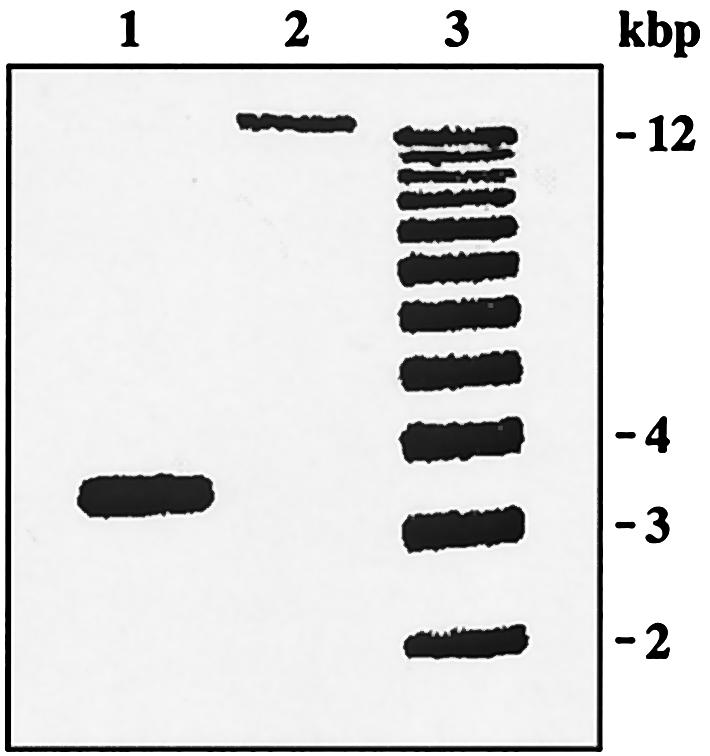
PCR amplification of fnr region from different MG1655 isolates. The fnr region was amplified from the CGSC isolate of MG1655 (CGSC 6300; lane 1) and the isolate obtained from M. Singer and C. Gross (NCM3430; lane 2) (see Materials and Methods). The sizes of the molecular standards in lane 3 are noted to the right. The genes deleted in the CGSC isolate (b1332 to b1344) are, respectively, ynaJ (open reading frame conserved in E. coli and Salmonella enterica), uspE (ydaA; UspA paralog) (17), fnr (Crp family activator of anaerobic respiratory gene transcription), ogt (O-6-alkylguanine-DNA/cysteine-protein methyltransferase), abgT (ydaH; p-aminobenzoyl-glutamate transport; oxidized folate recycling) (23), abgB (ydaI; p-aminobenzoyl-glutamate; oxidized folate recycling) (23), abgA (ydaJ; p-aminobenzoyl-glutamate; oxidized folate recycling) (23), abgR (ydaK; p-aminobenzoyl-glutamate regulator, LysR-type) (23), ydaL (open reading frame conserved in enterobacteria), ydaM (open reading frame conserved in E. coli), ydaN (open reading frame conserved in enterobacteria), dbpA (ATP-dependent RNA helicase), and ydaO (open reading frame conserved in enterobacteria). The deletion is flanked by tns5_4 (b1331), which codes for IS5 transposase, and ydaP (b1345), a rac prophage which codes for a putative prophage integrase.
