Figure 1.
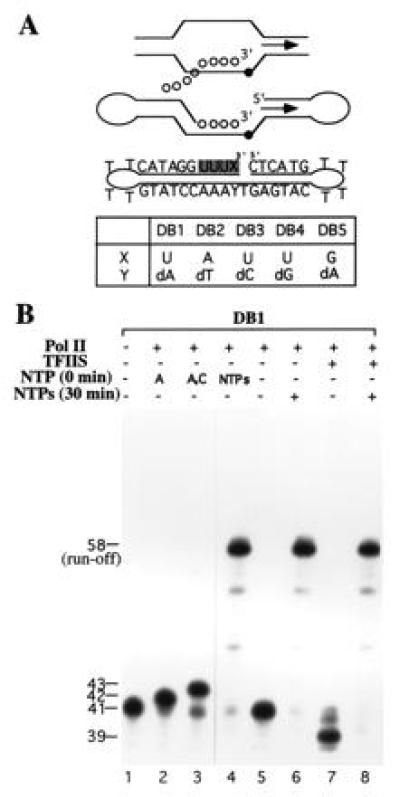
Architecture and transcription of RNA–DNA dumbbell by RNA pol II with and without TFIIS. (A) Schematics of the nucleic acid framework in the elongation bubble and its minimalistic mimic dumbbell. Arrows indicate transcription direction, and solid and open circles represent deoxy- and ribonucleotides, respectively. In the sequence of the dumbbell presented, the ribonucleotides are shaded. Several dumbbells in which the nature of the 3′-terminal ribonucleotide X and deoxynucleotide Y were varied are shown. (B) Elongation and cleavage of DB1 by pol II with and without TFIIS. RNA synthesis was carried out by pol II without preincubation (0 min) or after preincubation (30 min) with DB1 followed by addition of indicated NTP. The template-dependent elongation of the 41-nt dumbbell DB1, in the presence of four NTPs and pol II generates a run-off 58-nt RNA–DNA duplex. The ribonuclease activity of pol II cleaves DB1 into 39-nt dumbbell. In the RNA cleavage reaction, NTPs were omitted.
