FIG. 3.
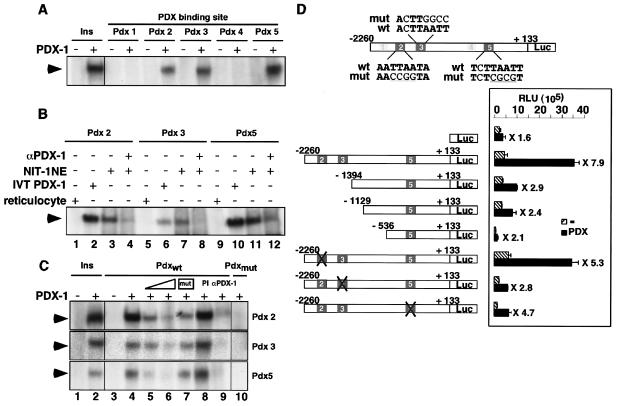
Human LRH-1 promoter is responsive to PDX-1 in vitro. (A) EMSA showing PDX-1 binding to three out of the five potential PDX-1 binding sites, Pdx 2, 3, and 5. Binding of PDX-1 to the PDX-1 consensus site in the insulin promoter (Ins) is shown as a positive control. (B) EMSA showing binding of in vitro-synthesized PDX-1 (IVT PDX-1, lanes 2, 6, and 10) and NIT-1 insulinoma nuclear extracts (NIT-1 NE, lanes 3, 7, and 11) to the Pdx 2, 3, and 5 sites of the human LRH-1 promoter. Incubation of NIT-1 nuclear extracts with an anti-PDX-1 antibody decreased the intensity of the retarded band, demonstrating the specificity of the binding (lanes 4, 8, and 12). No binding could be observed with nonprogrammed reticulocyte lysate (lanes 1, 5, and 9). (C) EMSA with the Pdx 2, 3, and 5 sites of the human LRH-1 promoter as probes. Increasing amounts of unlabeled wild-type oligonucleotides competed for binding with the radiolabeled Pdx 2, 3, and 5 oligonucleotides (lanes 5 and 6), whereas competition with the respective mutated oligonucleotides (mutations in sequences are indicated in panel D) did not displace the protein-DNA complex (lane 7). Incubation with a PDX-1 antibody abrogated the formation of the retarded band (α-PDX-1, lane 9), whereas no modification in PDX-1 binding was observed with a preimmune serum (PI, lane 8). No binding of PDX-1 to radiolabeled Pdx 2, 3s and 5 mutated oligonucleotides was observed (Pdxmut, lane 10). (D) Activity generated from the pGL3-LRH-1-2260 reporter cotransfected in 293T cells with an empty vector or an expression vector encoding murine PDX-1. Various unilateral deletion mutations as well as site-directed mutations in the putative PDX-1 sites of the human LRH-1 promoter indicated that the DNA region located between −2260 and −1394 confers responsiveness to PDX-1.