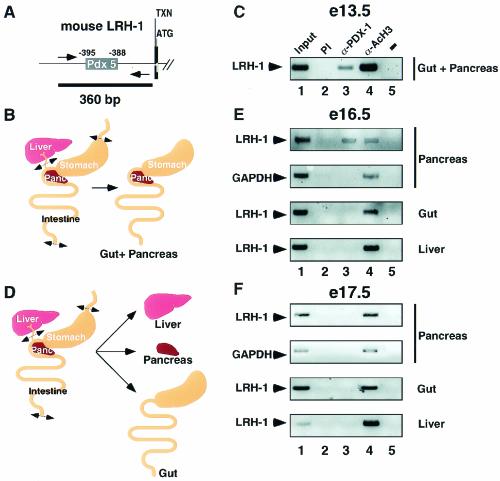
FIG. 5.
PDX-1 binds to the LRH-1 promoter during pancreatic development. (A) Schematic representation of the 5′ region of the mouse LRH-1 gene. The Pdx 5 binding site is indicated. Transcription (arrow, TXN) and translation (ATG) initiation sites are shown. Amplimers are highlighted by the arrows, whereas the black line depicts the 360-bp PCR product. (B) Scheme depicting the microdissection of the gut and pancreas used for the in vivo ChIP on E13.5 mouse embryos. (C) In vivo ChIP assay demonstrating binding of endogenous PDX-1 to the murine LRH-1 promoter in the gut and pancreas from E13.5 embryos. Chromatin from gut and pancreas was incubated with antibodies against PDX-1 (lane 3), acetylated histone H3 (lane 4), or preimmune serum (lane 2). Immunoprecipitates were analyzed by PCR with primers specific for the mouse LRH-1 promoter. A negative control with water as the template for PCR is shown in lane 5. Input chromatin was used as a positive control in lane 1. (D) Schematic representation showing the microdissection of the isolated organs used for in vivo ChIP on E16.5 (E) and E17.5 (F) embryos. (E) In vivo ChIP assay demonstrating occupancy of the murine LRH-1 promoter by PDX-1 in pancreas but not gut or liver of E16.5 embryos. Cross-linked chromatin from pancreas, gut, and liver was prepared for the ChIP assay as described for panel B. Immunoprecipitates were analyzed by PCR with primers specific for the murine LRH-1 and murine GAPDH (positive control) promoters. (F) PDX-1 does not occupy the LRH-1 promoter in endodermal tissues dissected from E17.5 mouse embryos. Cross-linked chromatin from the pancreas, gut, and liver of E17.5 embryos was analyzed by ChIP as described for panel D.