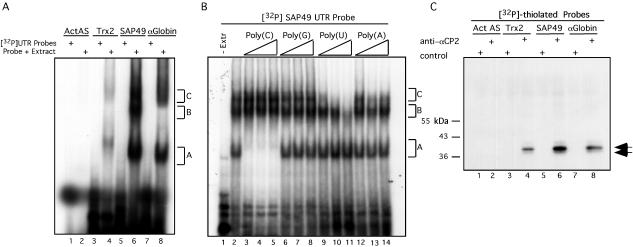
FIG. 5.
The 3′ UTRs of SAP49 and Trx2 mRNAs bind directly and specifically to αCP2. (A) RNA EMSA analysis. 32P-labeled RNAs corresponding to the four RNAs indicated at the top of the figure were incubated with K562 extract (Probe + Extract) or without K562 extract ([32P]UTR Probes) as indicated. Three observed RNP complexes are referred to as A, B, or C. β-Actin antisense (ActAS) and α-globin RNAs served as negative and positive controls, respectively. (B) EMSA analysis of the SAP49 3′ UTR in the presence of unlabeled homoribopolymer competitors. Triangles represent increasing amounts of competitor RNA used in the EMSA (10-, 100-, and 500-fold molar excess over the labeled probe). The competitor utilized is indicated above the triangles. (C) UV cross-linking of cytoplasmic proteins to RNA sequences. The same four RNA sequences analyzed by EMSA in Fig. 5A were UV cross-linked to K562 extract and immunoprecipitated with anti-αCP2 or control sera. Numbers on the left represent the migration of marker proteins. The arrows to the right of the autoradiograph indicate the cross-linked products representing αCP2 and αCP2-KL.