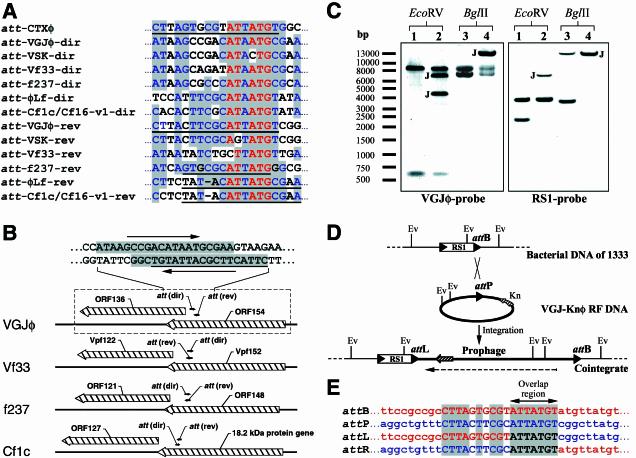
FIG. 2.
att site-containing region of VGJφ and site-specific integration of VGJ-Knφ into chromosome I of strain 1333. (A) Sequence alignment of the att regions of the CTXφ (accession number AF220606), VGJφ (AY242528), and VSK (NC_003327) phages of V. cholerae, the Vf33 (AB017573) and f237 (NC_002362) phages of V. parahaemolyticus, and the Cf1c (NC_001396), Cf16-v1 (M23621), and φLf (X70328) phages of X. campestris. Levels of identity greater than 90% are indicated by red letters with a gray background, and levels of identity greater than 45% are indicated by blue letters with a gray background. The underlined sequences indicate known functional att sites of the phages. (B) Genomic region containing the att-like sites shown in panel A. In all cases att sites map inside VGJφ ORF154 homologues, near VGJφ ORF136 homologues, which partially overlap ORF154 homologues. The region enclosed in a box (dashed lines) is the region enclosed in a box in Fig. 1A. (C) Southern blot showing the cointegrate structure in 1333/VGJK2, a representative clone. The left panel was probed with the VGJφ-specific probe (lanes 1 and 3, purified RF DNA of VGJ-Knφ; lanes 2 and 4, total DNA of the cointegrate clone). The right panel was hybridized with the RS1-specific probe (lanes 1 and 3, total DNA of strain 1333; lanes 2 and 4, total DNA of the cointegrate clone). DNA samples were treated with the enzymes indicated at the top. Bands containing the novel junctions are indicated by J. Note that the 3.5-kb BglII fragment of 1333 (right panel, lane 3) was about 9.1 kb larger in 1333/VGJK2 (double band in lane 4 of the right panel), indicating that only one copy of VGJ-Knφ was integrated. (D) Schematic representation of the recombination between the VGJ-Knφ RF (thick line) and the bacterial chromosome of 1333 (thin line). The RS1 element is represented by an open box flanked by the nearly identical end repeats (solid arrowheads). The downstream end repeat serves as an attB site for integration of VGJ-Knφ. attP (att-VGJφ-rev) and the novel junction attL and attR sites are also represented by solid arrowheads. Ev, EcoRV sites involved in the analysis of the Southern blot (panel A). The Knr gene (Kn) is represented by a cross-hatched arrow. The dashed arrow indicates the direction of the plus strand of VGJφ. (E) DNA region with attP, attB, attL, and attR sites as determined by sequencing. The core homologous sequences are indicated by uppercase letters, and identical nucleotides are indicated by a gray background. DNA sequences of bacterial origin and DNA sequences of phage origin are indicated by red and blue letters, respectively; black letters indicate sequences of unknown origin at the moment of recombination. The arrow indicates the putative overlap region, where the cutting and rejoining of the recombination partners took place.