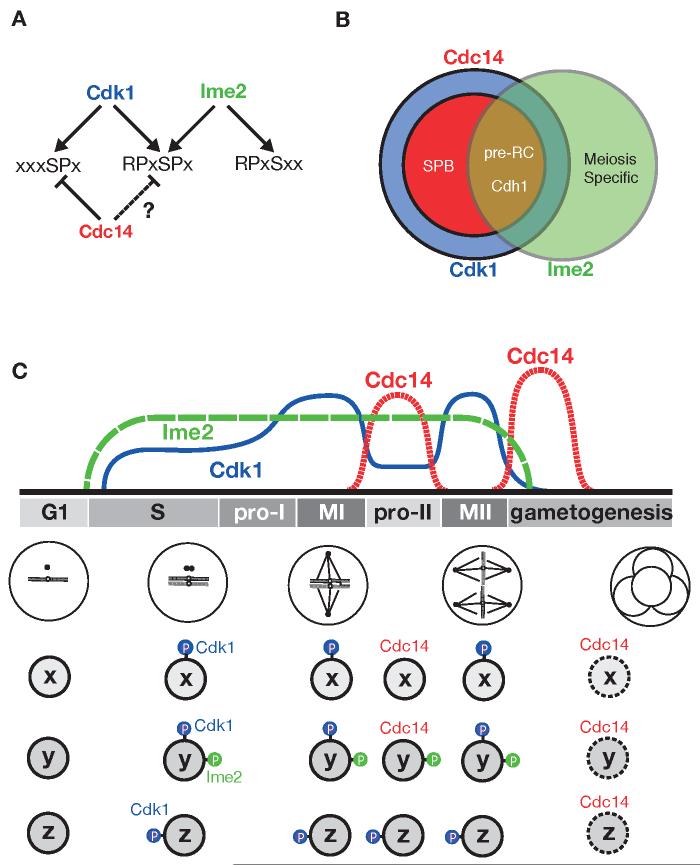
Figure 6.
A model for the phosphoregulation of the meiotic divisions
(A) Cdk1 sites (left) are sensitive to Cdc14 dephosphorylation; dual-specificity sites (middle) can be phosphorylated by both Cdk1 and Ime2 but their Cdc14 sensitivity is not known; and Ime2 sites (right) are resistant to dephosphorylation by Cdc14.
(B) The Cdk1 substrate set is represented by a blue circle, within which Cdc14 targets are represented as a red subset. Ime2 substrates are represented by a green circle. Ime2 may be specialized to regulate some processes unique to meiosis, such as recombination and gametogenesis, but also phosphorylates a subset of Cdk1 substrates. In the absence of Ime2, Cdc14 dephosphorylates a subset of Cdk1 substrates, including pre-RC components and Cdh1. In the presence of Ime2, however, we propose that Cdc14-resistant phosphates are placed on these and other substrates, helping to convert the single mitotic division into two consecutive meiotic divisions.
(C) The activities of Cdk1 (blue), Ime2 (green) and Cdc14 (red) are thought to rise and fall as shown during the meiotic program. Three hypothetical phosphoproteins are illustrated: protein x, a Cdk1 substrate that is dephosphorylated by Cdc14 between the meiotic divisions; protein y, a substrate for both Cdk1 and Ime2 whose Cdk1 sites, but not Ime2 sites, are dephosphorylated by Cdc14; and protein z, a Cdk1 substrate that remains phosphorylated between the meiotic divisions. Clb1-Cdk1, together with Ime2, drives the cell into meiosis I. Cdc14 is activated at the first metaphase-to-anaphase transition and removes Cdk1 phosphates (blue) from protein x and protein y. Protein x is thus dephosphorylated and activated (e.g. proteins involved in spindle cycle progression). However, the presence of an Ime2 phosphate (green) on protein y or a stable Cdk1 phosphate (blue) on protein z prevents them from being activated. We speculate that maintenance of phosphorylation on these proteins prevents origin licensing and resetting of the cell to a G1 state.