Figure 7.
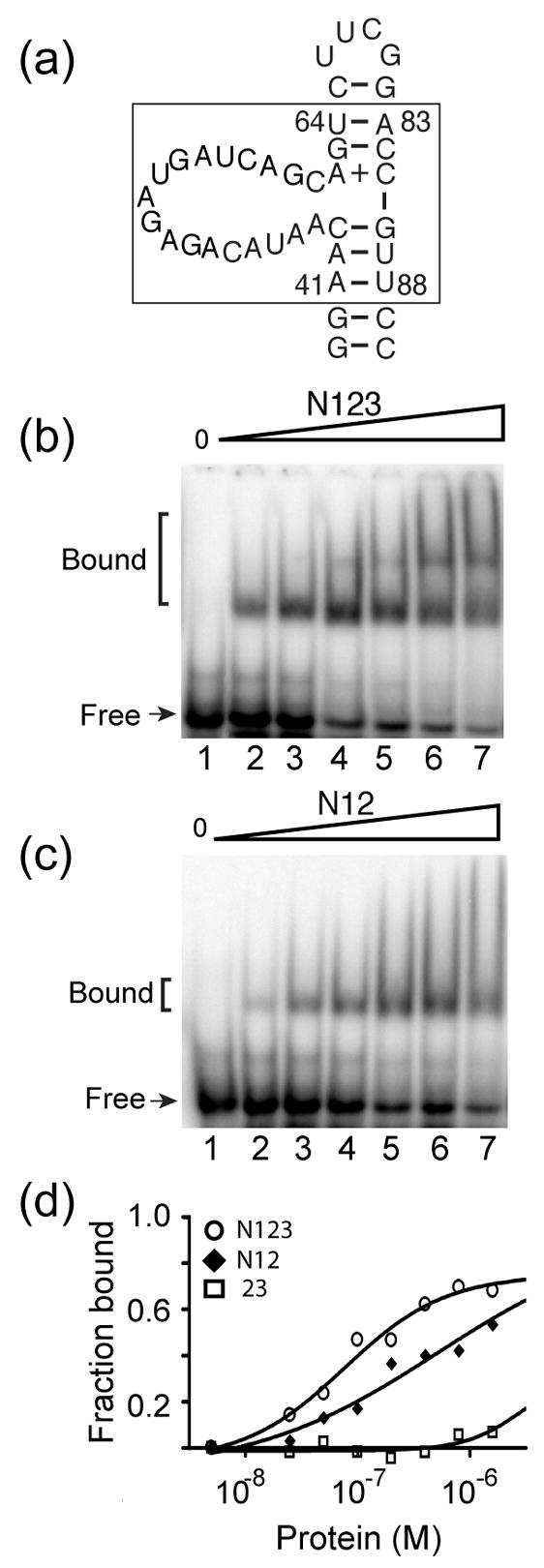
Binding of truncated Prp24 proteins to a domain of U6 RNA. (a) Primary and possible secondary structure of a minimal U6 RNA construct (40-nucleotide RNA) used in gel mobility shift assays (S. cerevisiae nucleotides 41–64 and 83–88 are boxed). 15N-labeled (b) Prp24-N123 and (c) Prp24-N12 proteins at concentrations of 0, 25, 50, 100, 200, 400, and 800 nM (lanes 1–7, respectively) were incubated with 32P-labeled 40-nucleotide RNA and resolved on a 6% native polyacrylamide gel. (d) Fraction of RNA bound plotted against the total concentration of Prp24-N123 (open circles), N12 (filled diamonds), and 23 (open squares). Data were fit to a one-site hyperbolic binding function (Y= (Bmax*X)/{Kd +X}), where Y is the fraction bound and X is the concentration of Prp24. The apparent Kd (in nM) is 45 ± 10 for Prp24-N123 and 81 ± 10 for Prp24-N12. An accurate apparent Kd value could not be extrapolated for Prp24-23.
