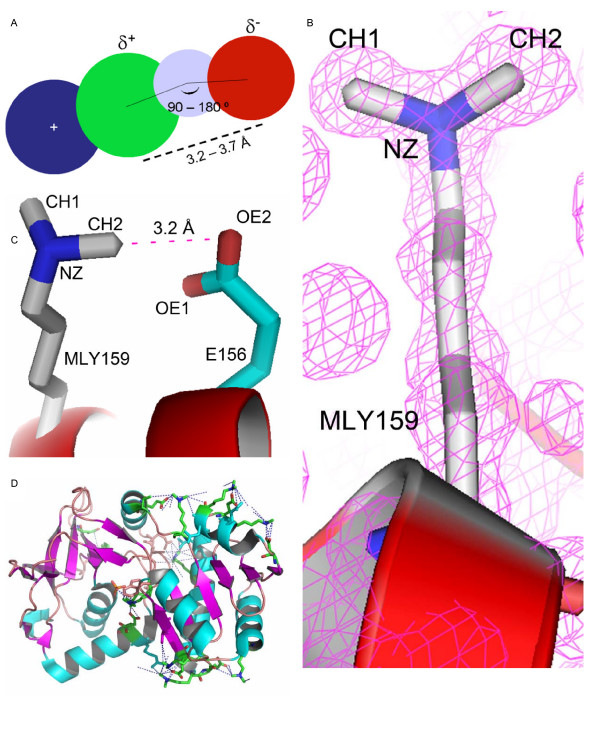
Figure 1.
Nature of (NZ)CH...O contacts of the methylated nuclease. A. Diagrammatic representation of the (NZ)CH...O bond. The side chain amine nitrogen (NZ) of lysine residues (blue circle) polarizes the covalently linked methyl carbon (green circle). The polarized methyl carbon acts like a proton donor and forms ionic interactions with neighbouring carboxyl oxygens (red circle). The optimal range for the (NZ)CH...O bond distance is between 3.2 – 3.7 Å. The angle of the approach of a proton towards the lone pair of electrons is generally between 90 – 180°. In order to calculate the angle, the position of hydrogen (grey circle) for X-ray structures is usually deduced. B. Electron density for the dimethylated lysine MLY159. The 2 | Fo | – | Fc | electron density map was contoured at 1.5 σ. C. The protonated methyl carbon, CH2, of MLY159 is seen forming a 3.6 Å (NZ)CH...O bond with the carboxyl oxygen of E156. The amino acids are represented as sticks. The (NZ)CH...O bond is shown as a dashed magenta line. D. A cartoon representation of the modified protein showing the dimethylated lysines engaged in the formation of numerous intra molecular (NZ)CH...O contacts. Such contacts localize side chains and loops in space, resulting in a compact protein molecule. (NZ)CH...O bonds are shown as dashed blue lines.