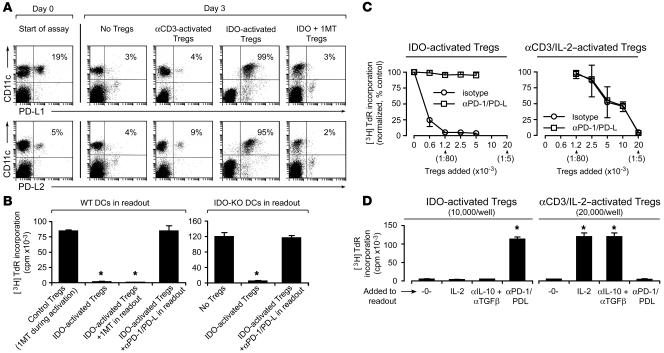
Figure 3. Suppression by IDO-activated Tregs requires the PD-1/PD-L pathway.
(A) Tregs were activated with IDO+ pDCs as described in Figure 2, then 1 × 104 sorted Tregs were added to readout assays (A1 T cells + CBA DCs). After 24 hours, cultures were harvested and stained for PD-L1 and PD-L2 relative to CD11c. Percentages indicate the proportion of cells that are dual-positive (right-upper quadrant). One of 3 experiments. (B) IDO-activated Tregs (5,000/well) were added to readout assays (A1 T cells plus either wild-type CBA DCs or IDO-KO DCs on the CBA background). Readout assays received either no additive, 1MT, or a cocktail of blocking antibodies against PD-1, PD-L1, and PD-L2 (50 μg/ml each). Control Tregs received 1MT during the activation step. One of 3 experiments; *P < 0.01 by ANOVA. (C) Tregs were activated with IDO+ pDCs or in identical cultures containing 1MT to block IDO and αCD3 plus IL-2 to activate the Tregs. After sorting, Tregs were added to readout assays (A1 T cells + CBA DCs) with or without PD-1/PD-L–blocking antibodies as shown. Graphs show the mean ± SD of 10 independent experiments with IDO-activated Tregs and 3 experiments with αCD3-activated Tregs, using TDLN pDCs from B78H1–GM-CSF and B16-OVA tumors. (D) IDO-activated Tregs (1 × 104/well) and αCD3/IL-2–activated Tregs (2 × 104/well) were prepared as described in the previous panel and added to readout assays with or without recombinant IL-2, anti–IL-10 plus anti–TGF-β blocking antibodies (100 μg/ml each), PD-1/PD-L–blocking antibodies, or no additive (-0-). Error bars show SD for replicate wells in 1 of 4 similar experiments. *P < 0.01 by ANOVA.