Abstract
Prenylated Rab GTPases regulate intracellular vesicle trafficking in eukaryotic cells by associating with specific membranes and recruiting a multitude of Rab-specific effector proteins. Prenylation, membrane delivery, and recycling of all 60 members of the Rab GTPase family are regulated by two related molecules, Rab escort protein (REP) and GDP dissociation inhibitor (GDI). Biophysical analysis of the interaction of prenylated proteins is complicated by their low solubility in aqueous solutions. Here, we used expressed protein ligation to construct a semisynthetic fluorescent analogue of prenylated Rab7, Rab7-NBD-farnesyl. This molecule is soluble in the absence of detergent but is otherwise similar in its behavior to naturally prenylated Rab7 GTPase. To obtain information on the interaction of natively mono- and diprenylated Rab7 GTPases with REP and GDI molecules, we stabilized the former molecules in solution by using the β-subunit of Rab geranylgeranyl transferase, which we demonstrate to function as an unspecific chaperone of prenylated proteins. Using competitive titrations of mixtures of natively prenylated and fluorescent Rab, we demonstrate that monogeranylgeranylated Rab7 binds to the REP protein with a Kd value of ≈70 pM. The affinity of doubly prenylated Rab7 is ≈20-fold weaker. In contrast, GDI binds both prenylated forms of Rab7 with comparable affinities (Kd = 1–5 nM) but has extremely low affinity to unprenylated Rab molecules. The obtained data allow us to formulate a thermodynamic model for the interaction of RabGTPases with their regulators and membranes and to explain the need for both REP and GDI in Rab function.
Keywords: geranylgeranyl, protein prenylation
Posttranslational prenylation of proteins with isoprenoid lipids is one of the most widespread and well recognized posttranslational modification of proteins in eukaryotic cells (1). In protein prenylation, either a 15-carbon-long farnesyl or a 20-carbon-long geranylgeranyl (GG) chain is donated from a soluble phosphoisoprenoid and enzymatically conjugated with C-terminal cysteine residues. The reaction is carried out by one of three structurally and functionally related protein prenyltransferases: farnesyltransferase (FTase), geranylgeranyltransferase-I (GGTase-I), or Rab GGTase (RabGGTase or GGTase-II) (2). The latter enzyme stands quite apart from the first two enzymes both functionally and structurally. RabGGTase has very strict substrate preference and acts only on the members of the RabGTPase family, which play a central role in membrane trafficking in all eukaryotic cells (3). Unlike other prenyltransferases, RabGGTase does not recognize a short C-terminal sequence but requires the integrity of the catalytic core of RabGTPases. Although, like other prenyltransferases, RabGGTase is a heterodimer of α- and β-subunits, it requires an additional unique factor termed Rab escort protein (REP). REP is a multifunctional protein that recruits a newly synthesized Rab GTPase and presents it to the RabGGTase by binding to its α-subunit (4). After the addition of, in most cases, two GG moieties onto C-terminal cysteines of Rab, the catalytic ternary complex dissociates and the prenylated Rab:REP complex travels to the destined membrane organelle. Insertion of GG moieties into lipid bilayers ensures stable association of RabGTPases with membranes (5). Membrane-bound Rabs are activated by nucleotide exchange and mediate processes of vesicular transport, docking, and fusion. Eventually, Rab proteins are converted into the GDP bound form and become available for extraction by GDP dissociation inhibitor (GDI) (6). Similar to REP, GDI is a tightly packed molecule composed of two domains, the larger of which forms an extended protein:protein interface with the catalytic domain of the GTPase, whereas the smaller one harbors conjugated isoprenoids (7, 8). GDI is able to extract prenylated Rab proteins from membranes as well as mediate their reinsertion. The process of extraction is believed to be thermodynamically favored and can occur spontaneously, but additional factors were proposed to be involved in its regulation (9, 10). Remarkably, the structurally and functionally related REP is inefficient in extracting Rab proteins from membranes despite its high affinity for these molecules (11, 12).
Comparative structural and functional analysis of REP and GDI revealed that the Rab binding site of REP is more extensive than that of GDI and allows it to bind unprenylated RabGTPases with nanomolar affinity. In contrast, GDI binds unprenylated Rab proteins with micromolar or worse affinity, and prenylation of Rabs is strictly required for stable complex formation (12). Recently, we proposed a model that postulated that the affinity difference in the REP/GDI interaction with prenylated and unprenylated forms of Rab proteins determines and distinguishes their role in the Rab cycle (9). Although informally consistent, this model was based on very approximate estimates of the prenylated Rab affinities for GDI and REP because technical difficulties associated with handling prenylated proteins. Geranylgeranylated proteins aggregate in aqueous solutions and require the presence of detergents to compensate for the hydrophobicity of the lipid moieties. Classical experiments used low concentrations of detergent to keep prenylated Rab9 in solution and estimate its affinity to GDI by measuring the influence of the GDI concentration on the rate of nucleotide release from the former (13). This study concluded that prenylated Rab9 has a nanomolar affinity for GDI while also demonstrating that the concentration of the detergent strongly influences Rab:GDI interactions. The affinity of REP for prenyated Rab is still unknown, making it impossible to develop a quantitative and comparative model of REP/GDI-mediated Rab cycling. This shared problem is in the analysis of geranylgeranylated proteins and is exemplified, for instance, by Rho and RhoGDI interactions, for which no reliable affinity estimates are available. In the presented study we have developed a soluble semisynthetic sensor that mimics prenylated Rab GTPase and displays a large fluorescence enhancement upon interaction with REP and GDI. Using this sensor, we were able to determine the absolute affinities of the interaction of GDI and REP with prenylated Rab7 GTPase. The data obtained allow us to construct a thermodynamic model of the cyclical interaction of Rab proteins with intracellular membranes.
Results
Construction of Soluble Semisynthetic Rab7-NBD-Farnesyl.
To gain insight into the interaction of prenylated Rab GTPase with its regulatory proteins or membranes, two interconnected technical issues need to be addressed. First, an efficient and if possible time-resolved method for monitoring the interaction of prenylated Rab with REP and GDI needs to be devised. Secondly, a way needs to be found to apply this assay to monitor the interaction of prenylated Rab with REP and GDI in the absence of detergent, which is known to strongly influence the properties of the prenylated proteins (13).
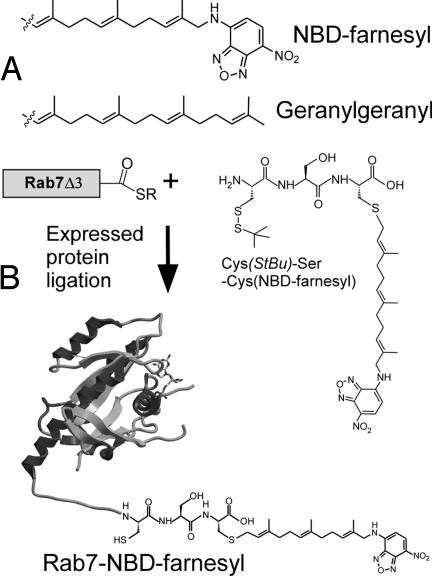
We recently developed an in vitro Rab prenylation assay that takes advantage of a 20-fold increase in the fluorescence intensity of a GG pyrophosphate (GGPP) analogue, 3,7,11-trimethyl-12-(7-nitro-benzo[1,2,5]oxadiazo-4-ylamino)-dodeca-2,6,10-trien-1 pyrophosphate (NBD-FPP) (Scheme Scheme 1A), upon its utilization as lipid substrate by RabGGTase (14). In the case of NBD-FPP, a monoprenylated reaction intermediate could dissociate from RabGGTase and needed to rebind to acquire the second prenyl moiety, in stark contrast to the native reaction (14, 15). This increased tendency to dissociate is a consequence of reduced hydrophobicity of the fluorescent analogue compared with the native isoprenoid. We conjectured that the NBD-farnesylated Rab molecule would on the one hand have the features of geranylgeranylated Rab proteins and on the other hand be more soluble than natively prenylated protein. To avoid technical difficulties associated with the large-scale enzymatic NBD-farnesylation, we chose an expressed protein ligation approach for construction of NBD-farnesylated Rab (16–18). To this end, we synthesized a tripeptide Cys-Ser-Cys(NBD-farnesyl) and ligated it in vitro to sodium 2-mercaptoethanesulfonate (MESNA) thioester-tagged Rab7 lacking three C-terminal amino acids (Scheme 1). The resulting protein remained in solution after removal of detergent and eluted from a size exclusion column at a position corresponding to a molecular mass <10 kDa whereas unligated protein eluted at the expected position corresponding to 28 kDa (Fig. 1A), suggesting that the protein does not form multimers and that its migration is retarded on the column probably by hydrophobic interaction with the matrix.
Scheme. 1.
Construction of prenylated fluorescent Rab7. (A) The fluorescent analogue of GG, NBD-farnesyl. (B) Synthesis of the fluorescent analogue of monogeranylgeranylated Rab, Rab7-NBD-farnesyl.
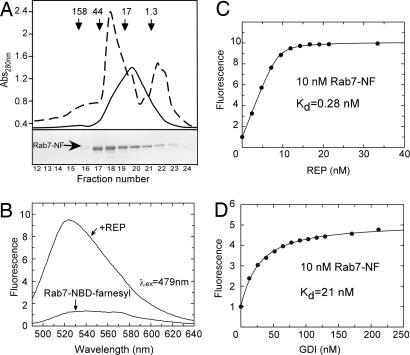
Fig. 1.
Gel filtration chromatography of semisynthetic Rab7-NF and analysis of its interaction with REP. (A) (Lower) Elution profile of Rab7-NF resolved on a Superdex 200 column and analysis of the resulting fractions by SDS/PAGE followed by Coomassie blue staining. (Upper) The dashed line represents absorbance at 280 nm, whereas the solid line represents fluorescence. The arrows in Upper indicate elution position of molecular mass standards in kilodaltons. Fractions 19–21 containing Rab7-NF at >80% purity were collected and used for further analysis (see SI Fig. 5A). (B) Emission spectra of Rab7-NF in the absence and in the presence of REP. (C) Titration of REP to a 10 nM solution of Rab7-NF. (D) Titration of GDI to a 10 nM solution of Rab7-NF. The fluorescence of NBD was excited at 479 nm, and the emission was collected at 525 nm. Kd values were obtained by fitting the data to the solution of a quadratic equation (see SI Appendix).
Semisynthetic Rab7-NBD-farnesyl (Rab7-NF) displayed the expected molecular mass [supporting information (SI) Fig. 5B] and was correctly folded as judged by its ability to form a stoichiometric complex with GDI (SI Fig. 6). The formation of the complex gives a first indication that the NBD-farnesyl group contributes to stabilization of the Rab:GDI interaction. To further assess the extent to which the semisynthetic Rab7-NF emulates the monoprenylated Rab protein we subjected it to in vitro prenylation by RabGGTase by using GG pyrophosphate (GGPP) as a substrate. In this case, the isoprenoid should be incorporated onto the C-terminal cysteine that was used as a ligation site (Scheme 1). The in vitro prenylation reaction was analyzed both by reversed phase HPLC (SI Fig. 5A) and MALDI-TOF (SI Fig. 5B), confirming that the GG group was conjugated to the previously free C-terminal cysteine. Therefore, the constructed semisynthetic protein closely mimics monoprenylated Rab7.
Analysis of the Rab7-NF Interaction with REP and GDI.
Because semisynthetic Rab7-NF is both soluble and active, we attempted to use the fluorescence of the conjugated NBD fluorophore to monitor its interaction with REP. As shown in Fig. 1B, addition of REP to a 30 nM solution of Rab7-NF resulted in a 10-fold increase of fluorescence and a blue shift from 545 to 525 nm. Similar behavior was observed upon addition of GDI, albeit with a 5-fold fluorescence increase (data not shown). The large fluorescence change enabled us to perform titration experiments to determine the affinity of the Rab7-NF:REP interaction, which was expected to be in the low nanomolar range and hence had to be assayed at very low concentrations of the reactants (19). Titration of 10 nM Rab7-NF with increasing concentrations of REP yielded a Kd value of 0.28 nM (Fig. 1C). A similar experiment using GDI yielded a Kd value of 21 nM (Fig. 1D). These data indicate that the Rab7-conjugated NBD-farnesyl group increases the affinity of Rab7:REP complex by a factor of ≈5 compared with unprenylated Rab (Kd = ≈1 nM; ref. 19), whereas its effect on the Rab7:GDI interaction is much more pronounced. Unprenylated RabGTPases interact with GDI with micromolar affinities implying that the presence of NBD-farnesyl increases the affinity of the complex by >100 fold (12).
Construction and Solubilization of Semisynthetic Geranylgeranylated Rab7.
The data described above provides indications of the different effects prenylation has on Rab:REP and Rab:GDI interactions. However, this system is artificial, and thus the data may be only an approximation of the native situation. Thus, we sought a way to repeat some of these experiments with natively geranylgeranylated Rab7. GG modified Rab proteins are soluble in the presence of detergents, however their presence significantly affects the interaction between prenylated Rab and GDI (13). Ideally, a chaperone would be needed that on the one hand would prevent aggregation of prenylated Rab but on the other hand not interfere with its protein:protein interactions. Because the hydrophobic nature of GG moieties is the primary cause for the insolubility of prenylated Rab, we speculated that RabGGTase, whose β-subunit contains a GG lipid-binding site, might function as an unspecific isoprenoid chaperone. To prevent interactions of RabGGTase with REP we chose to work with the β-subunit only (4).
In the light of this idea, semisynthetic monogeranylgeranylated Rab7 (Rab7-G) was prepared as described in Materials and Methods and was renatured in the presence of an equimolar amount of recombinant β-subunit of RabGGTase (βGGT). The mixture was subjected to size exclusion chromatography, which demonstrated that both proteins coeluted at a position corresponding to a molecular mass of ≈45 kDa (Fig. 2A). The native folding and functionality of stabilized monoprenylated Rab7 was assessed by in vitro prenylation and analyzed by SDS/PAGE in which the incorporation of NBD-farnesyl into Rab7-G was quantified (SI Fig. 7). The βGGT stabilized Rab7-G complex could be efficiently prenylated by RabGGTase, confirming that the protein was fully functional and that the presence of βGGT did not significantly influence its ability to interact with REP (SI Figs. 7 and 8). Prenylated Rab proteins could also be stabilized in solution by using delipidated BSA but required higher molar excess of the former (20).
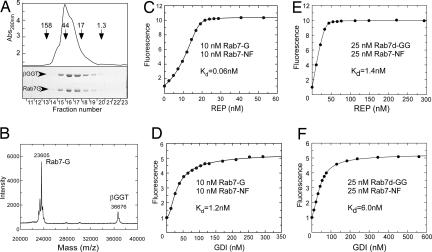
Fig. 2.
Solubilization of monogeranylgeranylated Rab7 by the β-subunit of RabGGTase and its interaction analysis with REP and GDI. (A) Gel filtration chromatography of Rab7-G:βGGT on a Superdex 200 column (Upper) and SDS/PAGE analysis of resulting fractions (Lower). (B) MALDI-TOF analysis of Rab7-G:βGGT complex (Mcalc = 23,660 Da for Rab7-G). (C and D) Titration of REP (C) or GDI (D) to a mixture of 10 nM Rab7-G:βGGT and 10 nM Rab7-NF. The data were fitted by numerical simulation and fitting to a competitive binding model to give Kd values for the interaction of Rab7-G with REP or GDI. (E and F) Titration of REP (E) or GDI (F) to a mixture of 25 nM Rab7-NF and 25 nM Rab7d-GG:BSA (λex/em:479/525 nm). The data were fitted to a competitive binding model to give Kd values of Rab7d-GG for REP and GDI, where Kd values of 0.22 nM and 21 nM of Rab7-NF for REP and GDI, respectively, were fixed.
Quantitative Analysis of the Interaction of Monogeranylgeranylated Rab7 with REP and GDI.
The availability of solubilized Rab7-G allowed us to devise an approach for monitoring its interaction with REP and GDI. Because Rab7-G by itself cannot be used as a reporter of the interaction, we decided to use the fluorescent Rab7-NF as a reporter. To this end, 10 nM Rab7-NF was mixed with 10 nM Rab7-G:βGGT complex, and the resulting solution was titrated with increasing concentrations of REP. As shown in Fig. 2C, there was an initial “lag” in the fluorescence increase, indicating that Rab7-G bound more strongly to the REP than Rab7-NF, initially resulting in the formation of a fluorescently silent complex. However, at higher REP concentrations, complex formation with Rab7-NF then occurred, leading to an increase in fluorescence intensity. Because the Kd value for the Rab7-NF:REP interaction had been determined independently, the data could be fitted numerically by using a competitive binding model leading to a Kd value of 61 ± 3 pM for the the Rab7-G:REP interaction. Rab7-NF binds βGGT with relatively low affinity (Kd = 114 ± 0.1 nM), and assuming a not dramatically higher affinity of Rab7-G, the influence of this interaction on the calculated Kd value for Rab7-G:REP interaction can be ignored (SI Figs. 9–13 and SI Tables 2–5). We applied the same approach to the analysis of the interaction of GDI with Rab7-G and determined a Kd value of 1.5 ± 0.3 nM for this interaction (Fig. 2D).
Interaction of Doubly Geranylgeranylated Rab7 with REP and GDI.
Natively monoprenylated Rab proteins comprise only a subgroup of mammalian RabGTPases and are absent in yeast. We therefore wished to explore the possible differences in the interaction of REP and GDI with the doubly and singly prenylated forms of Rab7. We previously reported synthesis of a semisynthetic doubly prenylated fluorescent variant, Rab7-A202C-E203K(dans)SC(GG)SC(GG), which we termed Rab7d-GG. The molecule displayed near-native properties and was used earlier as a fluorescent sensor to analyze the interaction of the prenylated Rab7:REP complex with RabGGTase (17).
Although obviously more hydrophobic, this molecule could be stabilized in solution in the absence of detergent with βGGT or delipidated BSA. Both approaches yielded native, correctly folded Rab7GTPase that could form a binary complex with REP as confirmed by gel filtration analysis (SI Fig. 14). To measure the affinities for these interactions, we used the above-described competitive titration with Rab7-NF in which a mixture of equimolar amount of Rab7-NF and Rab7d-GG:BSA was titrated with REP and GDI by using only the NBD fluorescence as a signal (Fig. 2 E and F). Remarkably, in this case the initial lag in fluorescence increase was not observed, already suggesting that diprenylated Rab7 binds to its regulators more weakly than its monoprenylated form. To confirm these data by using an independent signal, we took advantage of the dansyl group at position 203 that provides a sensitive fluorescence change upon interaction of Rab7d-GG with REP or GDI (SI Fig. 15 A and B) and that was used as a reporter in the titrations. Competitive titration of an equimolar mixture of Rab7d-GG and Rab7-G with REP by using the fluorescence of dansyl group, which again displayed a lag in the fluorescence change (decrease in this case) arising from preferential binding to the nonfluorescent monoprenylated form (SI Fig. 15 E and F). Numerical fit of the data from the competitive titrations of the Rab7-NF/Rab7d-GG and the Rab7d-GG/Rab7-G mixture with REP and GDI gave Kd values close to those obtained in direct titrations [Kd(Rab7d-GG:REP) = 1.3 nM, Kd(Rab7d-GG:GDI) = 5.0 nM] (see SI Figs. 15 C and D and 16 and Table 1).
Table 1.
Kd values of complexes of unprenylated wild-type Rab7 and differently prenylated Rab7 with REP and GDI
| Rab proteins | Kd for REP, nM | Kd for GDI, nM |
|---|---|---|
| Rab7wt | 1* | >5 × 104† |
| Rab7-NF | 0.22 ± 0.06 | 14 ± 5.5 |
| Monogeranylgeranylated Rab7 | 0.061 ± 0.03 | 1.5 ± 0.3 |
| Digeranylgeranylated Rab7 | 1.3 ± 0.2 | 5.2 ± 2.2 |
*See ref. 19.
†Determined by isothermal titration calorimetry (data not shown).
Discussion
In the presented work, we have used a combination of organic synthesis and expressed protein ligation to synthesize a fluorescent isoprenylated GTPase, Rab7-NF, that mimics monogeranylgeranylated Rab7. The protein was soluble and monomeric, in stark contrast to the behavior of geranylgeranylated proteins. Although the reduction in the length of the isoprenoid chain is known to decrease the hydrophobicity of the prenylated protein (21–23), it was previously not known to what extent these modifications affect isoprenoid mediated protein:protein interactions. The resulting protein displays features of a prenylated GTPase, such as the ability to form a stable high-affinity complex with GDI, which strictly requires Rab prenylation. This finding indicates that the semisynthetic GTPase closely mimics the natively prenylated Rab protein. Most importantly, Rab7-NF binding to REP or GDI results in a large fluorescence change, providing a highly sensitive readout for monitoring Rab-REP/GDI complex formation in real time. The dramatic enhancement in fluorescence is presumably a consequence of the Rab-conjugated NBD-farnesyl group binding in the hydrophobic lipid binding site on REP and GDI (11, 12, 14). Using this signal, we were able to obtain the affinities for REP/GDI interaction with prenylated proteins and show that the modification increased the affinity of Rab7 for REP by just by a factor of 4, whereas the affinity for GDI was increased by at least 1,000-fold.
To provide an independent confirmation to this conclusion, we performed the interaction analysis on natively prenylated Rab7 stabilized in solution by the recombinant β-subunit of RabGGTase. We find that the β-subunit binds protein-conjugated isoprenoids unspecifically with submicromolar affinity, which is not surprising considering the fact that prenyltransferases typically remain in complex with their products and that these are eventually released by the binding of new lipid substrate molecules (24, 25). We believe that the presented method will be applicable to all prenylated polypeptides and should enable quantitative analysis of their interactions with proteins and membranes.
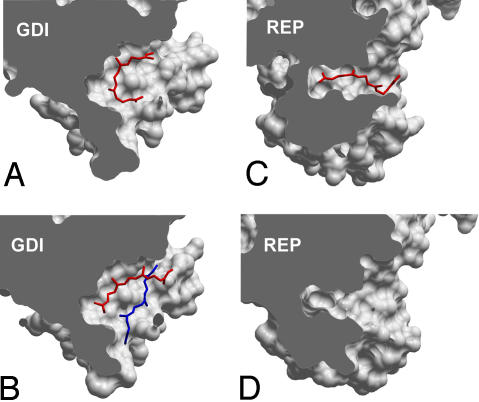
Analysis of the interactions of prenylated Rab7 with REP and GDI revealed that they display comparable affinities to the diprenylated form of this GTPase. In contrast, its monoprenylated form bound 3 times more tightly than the diprenylated form to GDI and nearly 20 times more tightly to REP. The latter observation finds support in earlier findings that, unlike the diprenylated protein, the monoprenylated Rab1 is not easily dissociated from REP by detergent and phospholipid, an observation that was interpreted to reflect a higher affinity of the monoprenylated Rab form for REP compared with diprenylated Rab (26). The small difference in the affinities of mono- and diprenylated Rab7 for GDI can be rationalized on the basis of the recently solved structures of Ypt1:GDI complexes (7, 12). In the structure of the monoprenylated Ypt1:GDI complex, the conjugated lipid is inserted into the lipid-binding pocket in a bent conformation, with both ends being solvent exposed, partially occupying the binding sites of both lipids (Fig. 3A). In the doubly prenylated Ypt1:GDI complex, one isoprenoid is buried at the bottom of the hydrophobic binding site, whereas the second stacks on top of it, forming relatively few contacts with GDI (Fig. 4B). Because of this situation, the overall contact area of the protein:lipid interface is increased only modestly upon conjugation of the second isoprenoid. The situation with the REP:Rab interaction appears to be more complex. Monoprenylation increases the affinity of the Rab:REP complex to an affinity of 60 pM, which is then reduced to ≈1 nM after the attachment of the second lipid. This observation implies that the prenyl groups bind only very weakly to REP in the diprenylated complex. This point was confirmed in the presented experiment (SI Fig 17). At the mechanistic level, these observations can be rationalized in the following way: The binding site of REP is narrower than that of GDI, and the lipid is inserted in an extended conformation (Fig. 3C). The structure of the doubly prenylated REP:Rab complex is not available, but should its lipid-binding site be already fully dilated in the complex with the monoprenyated GTPase, then the second lipid could bind outside of it (12). This situation may also lead to partial displacement of the isoprenoid moiety from the lipid-binding site, leading to the observed affinity decrease. The “purpose” of this mechanism may be the retention of the monoprenylated reaction intermediate in the complex to assure its complete processing. However, this model raises the question about the way natively monoprenylated RabGTPases are processed, because they may form a complex too tight to be dissociated for membrane insertion. Indirect support of this idea comes from the observation that monocysteine Rab5a and Rab27a mutants are retained on the ER membrane and are not delivered to their native locations (27). The answer probably lies in a large variation of affinities for the interaction of the Rab protein core with REP, with Rab7 being one of the tightest binders (11). With many Rab:REP interactions displaying Kd values of several hundred nanomolars, the overall affinity of monoprenylated complexes can be expected to remain in the nanomolar range.
Fig. 3.
Comparison of lipid binding sites of GDI and REP molecules in complex with prenylated Rab GTPases. (A) Monoprenylated Ypt1:GDI complex (1UKV). (B) Diprenylated Ypt1:GDI complex (2BCG). (C) Monoprenylated Rab7:REP complex (1VG0). (D) Unprenylated Rab7Δ22:REP complex (1VG9). The complexes were optimally superimposed, and the domains II were sliced to expose the lipid-binding site. All molecular manipulations including generation of images were performed with ICM Browser Pro (Molsoft, Redmond, WA).
Fig. 4.
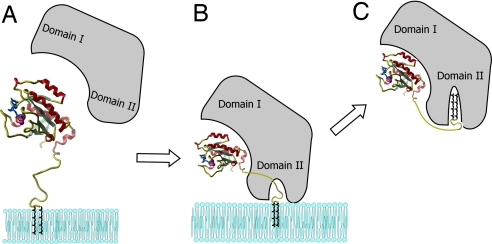
Model for the extraction of Rab from membranes by GDI or REP. (A) Initial recognition of the membrane associated Rab. (B) Formation of the membrane-bound Rab:REP/GDI complex. (C) Translocation of the lipid moiety to the GDI/REP and release form the membrane.
Finally, and probably most importantly, our study provides evidence for the recently proposed model of Rab GTPase membrane delivery and extraction (9). It was shown that REP appears to be much less efficient than GDI in Rab extraction. This difference in efficiency is in keeping with their biological roles, because REP is probably only involved in delivery of Rabs to membranes, whereas GDI, in addition to having this property, must also be able to extract them. These different properties can be explained by considering the affinities of REP/GDI for unmodified and prenylated forms of Rab, respectively. As shown in Table 1, whereas REP binds with the same high affinity to both unprenylated and diprenylated Rab, enabling it to present the unprenylated form to RabGGTase, GDI binds to prenylated Rab with at least 1,000-fold higher affinity than to unprenylated Rab. The large increase in affinity of GDI to Rab on docking of the C terminus and the conjugated isoprenoid groups is the driving force for the extraction process. Expressed in another manner, GDI is efficient in extraction of Rabs from membrane because there is a large difference in binding energy between the situations in which only the GTPase domain interacts with GDI and the situation in which the C terminus and conjugated isoprenoid moiety are also docked. Therefore, the difference in binding energies provides the thermodynamic driving force for the extraction from the membrane. In contrast, most of the binding energy in the case of REP comes from the interaction with the GTPase domain, with only very little or no driving force for the extraction provided by the interaction of the isoprenoid conjugated C terminus. As a consequence, REP has very low potential to extract Rab from membranes, but can readily release the prenyl groups for insertion into the membrane. A remaining problem concerns the dissociations of REP from a Rab molecule that has inserted its lipid into a membrane. On the basis of what is known about the kinetics of dissociation of REP from unprenylated Rab, this should occur at a rate of ≈0.01 s−1, corresponding to a half-life in the range of 1 min (19). It is possible that this is rapid enough for the physiological role of REP, especially when it is considered that after GDP to GTP exchange, a process which happens on the membrane, the affinity of REP for Rab is reduced by at least a factor of 10 through acceleration of the dissociation rate (19). However, it is possible and likely that additional proteinaceous factors, including GDI displacement factors (GDF), such as Yip proteins, are involved in this process, providing an additional level of regulation (28, 29).
Materials and Methods
Synthesis of Prenylated Peptides.
The synthesis of H2N-Cys(StBu)-Cys(GG)-OH and H2N-Cys(StBu)-Lys(dansyl)-Ser-Cys(GG)-Ser-Cys(GG)-OMe was described in refs. 7, 12, and 17. For the synthesis of H2N-Cys(StBu)-Ser-Cys(NBD-farnesyl)-OH, fluorescent lipid was first synthesized as described in ref. 30. The fluorescent lipid was coupled to cysteine followed by introduction of the Fmoc protecting group. The resulting Fmoc-Cys(NBD-farnesyl)-OH was coupled to 2-chlorotrityl chloride resin, and the tripeptide was synthesized by Fmoc solid-phase peptide synthesis strategy using N,N′-diisopropylcarbodiimide (DIC) and N-hydroxybenzotriazole (HOBt) as coupling reagents. The Fmoc group was removed using 20% piperidine in dimethylformamide (DMF). The tripeptide was released from the resin with 1% TFA and 1% TES in CH2Cl2 to give H2N-Cys(StBu)-Ser-Cys(NBD-farnesyl)-OH with a yield of 11%. For details of preparation, see SI Appendix. The mercapto group of the N-terminal cysteine side remained protected until the ligation reaction, during which in situ deprotection occurred due to excess of thiol reagent.
Protein Expression and Purification.
The Rab7 coding region truncated by 2-, 3-, or 6-aa residues was C-terminally fused to an intein-chitin binding domain assembly as implemented in the pTWIN-1 vector (New England Biolabs, Ipswich, MA). Protein expression in Escherichia coli and purification of thioester-tagged proteins was performed as described in ref. 17. The resulting Rab7-MESNA thioester protein was desalted on a PD-10 column (GE Healthcare, Piscataway, NJ) equilibrated with ligation buffer (25 mM Na-phosphate, pH 7.5/25 mM NaCl/2 mM MgCl2/10 μM GDP) and concentrated to 10–20 mg/ml.
Bovine α-RabGDI was expressed in SF9 cells by using the baculoviral expression system as a fusion with an N-terminal 6His tag and the tobacco etch virus (TEV) protease cleavage site. Protein was purified to homogeneity by a combination of Ni-NTA chromatography, proteolytic removal of the His tag and gel filtration as described for Rab7 GTPase (31). Recombinant REP-1 was expressed and purified as described in ref. 31. The β-subunit of rat RabGGTase (βGGT) was cloned into the pGATEV-mod vector where it is expressed as an N-terminal fusion with a 6His-GST assembly (32). The fusion protein was expressed in E. coli and purified to homogeneity by a combination of Ni-NTA chromatography, proteolytic removal of 6His-GST tag, and gel filtration as described for RabGGTase (32).
In Vitro Protein Ligation.
Preparation of the digeranylgeranylated Rab7 [Rab7-A202C-E203K(dans)SC(GG)SC(GG)] by in vitro protein ligation was performed as described in ref. 17.
Preparation of the monogeranylgeranylated Rab7 [Rab7-C205S-CC(GG)] by in vitro protein ligation was performed essentially the same as for monogeranylgeranylated Ypt1 as described in ref. 7, in which a thioester-tagged Rab7Δ2C205S was ligated with the peptide Cys(StBu)-Cys(GG).
To prepare Rab7CSC(NBD-farnesyl), Rab7Δ3-MESNA thioester protein (>10 mg/ml) in ligation buffer was supplemented with 40 mM cetyltrimethylammonium bromide (CTAB) and 100 mM MESNA. Ligation was initiated by adding >2 mM peptide from a ≈40 mM stock solution in DMSO. The reaction mixture was incubated overnight at 40°C with vigorous agitation. The reaction mixture was centrifuged, and the supernatant was removed. The pellet was washed four times with methanol, four times with of dichloromethane, four times with methanol, and four times with Milli-Q water at room temperature to remove untreated peptide and unligated protein. The precipitate was dissolved in buffer A [100 mM Tris·HCl, pH 8.0/6 M guanidinium-HCl/100 mM dithioerythritol (DTE)/1% CHAPS/1 mM EDTA] to a final protein concentration of 0.5–1.0 mg/ml and incubated overnight at 4°C with slight agitation. The solution was cleared by centrifugation.
Preparation of the Mono- and Diprenylated Rab7 Complexed to βGGT.
Mono- or digeranylgeranylated Rab7 in denaturation buffer was renatured by diluting it at least 30-fold drop-wise into refolding buffer (50 mM Hepes, pH 7.5/2.5 mM DTE/2 mM MgCl2/100 μM GDP/1% CHAPS/400 mM arginine-HCl/400 mM trehalose/1 mM PMSF) in the presence of an equimolar amount of βGGT with gentle stirring at room temperature. Alternatively, 10 molar excess of delipidated BSA was used. The mixture was incubated for 30 min at room temperature and 60 min on ice and concentrated to 2–5 mg/ml by using size exclusion concentrators (molecular mass cutoff, 10 kDa). The concentrated mixture was dialyzed overnight against two 2-liter changes of buffer B [25 mM Hepes, pH 7.5/50 mM (NH4)2SO4/50 mM NaCl/2 mM MgCl2/2.5 mM DTE/10 μM GDP/10% glycerol/1 mM PMSF]. The dialyzed material was centrifuged to remove aggregates and subsequently loaded on a Superdex-200 gel filtration column (GE Healthcare) equilibrated with buffer C (50 mM Hepes, pH 7.2/50 mM NaCl/5 mM DTE/2 mM MgCl2/10 μM GDP). The fractions containing the Rab7:βGGT complex were collected and concentrated to 2 mg/ml and were stored frozen at −80°C. Rab7CSC(NBD-farnesyl) was constructed, refolded, and purified as described above, except that βGGT was omitted.
Fluorescence Measurements.
Fluorescence spectra and long-time base-fluorescence measurements were performed with a Spex Fluoromax-3 spectrofluorometer (Jobin Yvon, Edison, NJ). All reactions were followed at 25°C in 50 mM Hepes pH 7.2, 50 mM NaCl, 2 mM MgCl2, and 5 mM DTE in a volume of 1 ml. Data analysis was performed with the program Grafit 5.0 (Erithacus software; Erithacus, Surrey, U.K.) and Scientist (MicroMath Scientific software; MicroMath, Salt Lake City, UT) as described in ref. 33.
In Vitro Prenylation Assay.
At room temperature, 6 μM Rab7-NF or Rab7-G, 6 μM REP, 6 μM RabGGTase, and 40 μM GG pyrophosphate (GGPP) or 3,7,11-trimethyl-12-(7-nitro-benzo[1,2,5]oxadiazo-4-ylamino)-dodeca-2,6,10-trien-1 pyrophosphate (NBD-FPP) were incubated for 30 min. In the control reactions, REP were omitted. The reactions were quenched by the addition of 0.1% trifluoroacetic acid, and the sample was subsequently subjected to HPLC, SDS/PAGE, electrospray ionization (ESI)-MS, and MALDI-TOF analysis. HPLC analysis was performed on a C4, 150 × 4.6 mm, 15 μm LUNA column (Phenomenx, Torrance, CA) driven by a Waters (Milford, MA) 600s system. The column was equilibrated with 95% buffer D (0.1% trifluoroacetic acid in water) and 5% buffer E (0.1% trifluoroacetic acid in 100% acetonitrile) at a flow rate of 1 ml/min. After injection and a 2-min wash step, the column was eluted with a gradient from 5% to 70% of buffer E in 15 min, followed by elution for 8 min with 70% buffer E. Liquid chromatography (LC)-ESI-MS analysis was performed on an Agilent 1100 series chromatography system (Hewlett–Packard, Palo Alto, CA) equipped with an LCQ electrospray mass spectrometer (Finnigan, San Jose, CA). MALDI spectra were recorded on a Voyager-DE Pro Biospectrometry workstation from Applied Biosystems (Weiterstadt, Germany). SDS/PAGE analysis of NBD-farnesylated proteins was performed using the Fluorescent Image Reader FLA-5000 (Fuji, Tokyo, Japan) as described in ref. 34.
Supplementary Material
Acknowledgments
We thank Dr. T. Durek for advice and for help at the initial stages of the project. K.A. was supported by a Heisenberg Award of the Deutsche Forschungsgemeinschaft. This work was supported in part by Deutsche Forschungsgemeinschaft Grants AL 484/7-2 (to K.A.) and SFB642 (to K.A., R.S.G, and H.W.).
Abbreviations
- βGGT
β-subunit of RabGGTase
- DTE
dithioerythritol
- GDI
GDP dissociation inhibitor
- GG
geranlygeranyl
- GGTase
geranylgeranyltransferase
- MESNA
sodium 2-mercaptoethanesulfonate
- NBD
3,7,11-trimethyl-12-(7-nitro-benzo[1,2,5]oxadiazo-4-ylamino)
- RabGGTase
Rab GGTase
- REP
Rab escort protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701817104/DC1.
References
- 1.Gelb MH. Science. 1997;275:1750–1751. doi: 10.1126/science.275.5307.1750. [DOI] [PubMed] [Google Scholar]
- 2.Casey PJ, Seabra MC. J Biol Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 3.Stenmark H, Olkkonen VM. Genome Biol. 2001;5:3007.1–3007.7. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pylypenko O, Rak A, Reents R, Niculae A, Sidorovitch V, Cioaca MD, Bessolitsyna E, Thoma NH, Waldmann H, Schlichting I, et al. Mol Cell. 2003;11:483–494. doi: 10.1016/s1097-2765(03)00044-3. [DOI] [PubMed] [Google Scholar]
- 5.Ghomashchi F, Zhang X, Liu L, Gelb MH. Biochemistry. 1995;34:11910–11918. doi: 10.1021/bi00037a032. [DOI] [PubMed] [Google Scholar]
- 6.Araki S, Kikuchi A, Hata Y, Isomura M, Takai Y. J Biol Chem. 1990;265:13007–13015. [PubMed] [Google Scholar]
- 7.Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 8.Schalk I, Zeng K, Wu SK, Stura EA, Matteson J, Huang M, Tandon A, Wilson IA, Balch WE. Nature. 1996;381:42–48. doi: 10.1038/381042a0. [DOI] [PubMed] [Google Scholar]
- 9.Goody RS, Rak A, Alexandrov K. Cell Mol Life Sci. 2005;62:1657–1670. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CY, Balch WE. Mol Biol Cell. 2006;17:3494–3507. doi: 10.1091/mbc.E05-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rak A, Pylypenko O, Niculae A, Pyatkov K, Goody RS, Alexandrov K. Cell. 2004;117:749–760. doi: 10.1016/j.cell.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Pylypenko O, Rak A, Durek T, Kushnir S, Dursina BE, Thomae NH, Constantinescu AT, Brunsveld L, Watzke A, Waldmann H, et al. EMBO J. 2006;25:13–23. doi: 10.1038/sj.emboj.7600921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro AD, Pfeffer SR. J Biol Chem. 1995;270:11085–11090. doi: 10.1074/jbc.270.19.11085. [DOI] [PubMed] [Google Scholar]
- 14.Wu YW, Waldmann H, Reents R, Ebetino FH, Goody RS, Alexandrov K. Chembiochem. 2006;7(12):1859–1861. doi: 10.1002/cbic.200600377. [DOI] [PubMed] [Google Scholar]
- 15.Thoma NH, Niculae A, Goody RS, Alexandrov K. J Biol Chem. 2001;276:48631–48636. doi: 10.1074/jbc.M106470200. [DOI] [PubMed] [Google Scholar]
- 16.Alexandrov K, Heinemann I, Durek T, Sidorovitch V, Goody RS, Waldmann H. J Am Chem Soc. 2002;124:5648–5649. doi: 10.1021/ja017799e. [DOI] [PubMed] [Google Scholar]
- 17.Durek T, Alexandrov K, Goody RS, Hildebrand A, Heinemann I, Waldmann H. J Am Chem Soc. 2004;126:16368–16378. doi: 10.1021/ja046164n. [DOI] [PubMed] [Google Scholar]
- 18.Muir TW. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 19.Alexandrov K, Simon I, Iakovenko A, Holz B, Goody RS, Scheidig AJ. FEBS Lett. 1998;425:460–464. doi: 10.1016/s0014-5793(98)00290-7. [DOI] [PubMed] [Google Scholar]
- 20.Dirac-Svejstrup AB, Soldati T, Shapiro AD, Pfeffer SR. J Biol Chem. 1994;269:15427–15430. [PubMed] [Google Scholar]
- 21.Dursina BE, Reents R, Niculae A, Veligodsky A, Breitling R, Pyatkov K, Waldmann H, Goody RS, Alexandrov K. Protein Expr Purif. 2005;39:71–81. doi: 10.1016/j.pep.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Chehade KA, Kiegiel K, Isaacs RJ, Pickett JS, Bowers KE, Fierke CA, Andres DA, Spielmann HP. J Am Chem Soc. 2002;124:8206–8219. doi: 10.1021/ja0124717. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen UT, Cramer J, Gomis J, Reents R, Gutierrez-Rodriguez M, Goody RS, Alexandrov K, Waldmann H. ChemBioChem. 2007;8:408–423. doi: 10.1002/cbic.200600440. [DOI] [PubMed] [Google Scholar]
- 24.Tschantz WR, Furfine ES, Casey PJ. J Biol Chem. 1997;272:9989–9993. doi: 10.1074/jbc.272.15.9989. [DOI] [PubMed] [Google Scholar]
- 25.Thoma NH, Iakovenko A, Kalinin A, Waldmann H, Goody RS, Alexandrov K. Biochemistry. 2001;40:268–274. doi: 10.1021/bi002034p. [DOI] [PubMed] [Google Scholar]
- 26.Shen F, Seabra MC. J Biol Chem. 1996;271:3692–3698. doi: 10.1074/jbc.271.7.3692. [DOI] [PubMed] [Google Scholar]
- 27.Gomes AQ, Ali BR, Ramalho JS, Godfrey RF, Barral DC, Hume AN, Seabra MC. Mol Biol Cell. 2003;14:1882–1899. doi: 10.1091/mbc.E02-10-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivars U, Aivazian D, Pfeffer SR. Nature. 2003;425:856–859. doi: 10.1038/nature02057. [DOI] [PubMed] [Google Scholar]
- 29.Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR. EMBO J. 1997;16:465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reents R, Wagner M, Schlummer S, Kuhlmann J, Waldmann H. ChemBioChem. 2005;6:86–94. doi: 10.1002/cbic.200400233. [DOI] [PubMed] [Google Scholar]
- 31.Alexandrov K, Simon I, Yurchenko V, Iakovenko A, Rostkova E, Scheidig AJ, Goody RS. Eur J Biochem. 1999;265:160–170. doi: 10.1046/j.1432-1327.1999.00699.x. [DOI] [PubMed] [Google Scholar]
- 32.Kalinin A, Thoma NH, Iakovenko A, Heinemann I, Rostkova E, Constantinescu AT, Alexandrov K. Protein Expr Purif. 2001;22:84–91. doi: 10.1006/prep.2001.1423. [DOI] [PubMed] [Google Scholar]
- 33.Alexandrov K, Scheidig AJ, Goody RS. Methods Enzymol. 2001;329:14–31. doi: 10.1016/s0076-6879(01)29062-4. [DOI] [PubMed] [Google Scholar]
- 34.Dursina B, Reents R, Delon C, Wu Y, Kulharia M, Thutewohl M, Veligodsky A, Kalinin A, Evstifeev V, Ciobanu D, et al. J Am Chem Soc. 2006;128:2822–2835. doi: 10.1021/ja052196e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.