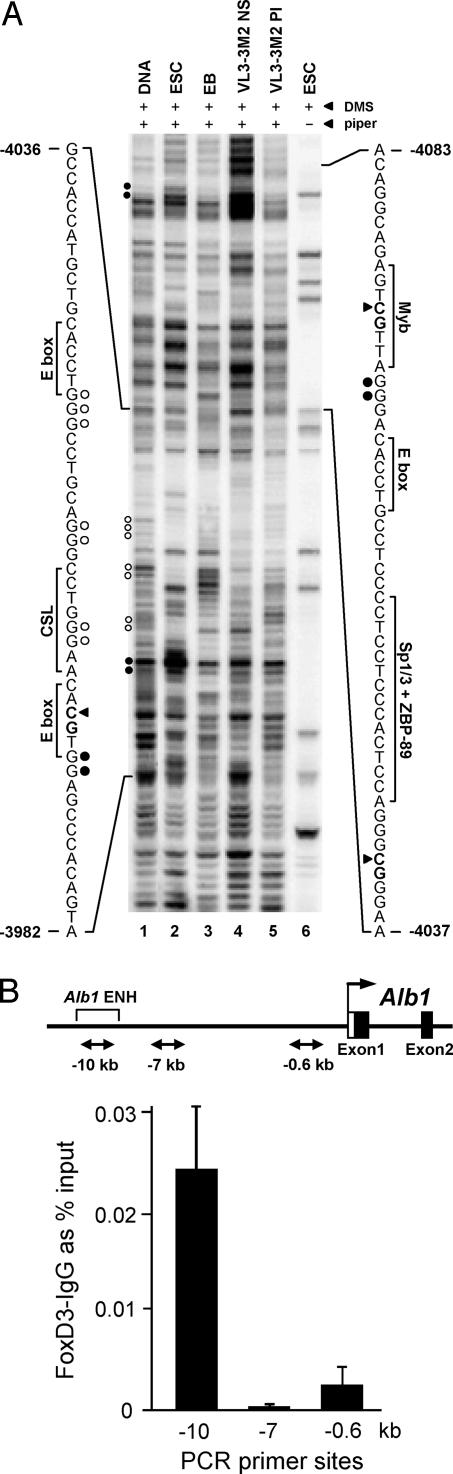
Fig. 4.
Binding of transcription factors to the Ptcra and Alb1 enhancers in ES cells. (A) In vivo protein–DNA interactions at the Ptcra enhancer region. DMS footprinting was performed with purified CCE ES cell DNA (lane 1), CCE ES cells (lane 2), EB cells (lane 3), Ptcra+ VL3-3M2 thymocytes (lane 4), Ptcra− PMA/ionomycin-treated VL3-3M2 cells (lane 5), and CCE ES cells without piperidine cleavage of methylated DNA (lane 6). Nucleotides consistently protected from methylation in ES cells (open circles) and nucleotides exhibiting enhanced methylation (filled circles) are shown. Potential transcription factor binding sites are shown. Filled triangles mark the positions of CpGs (−3,997, −4,042, and −4,080). SI Fig. 10 shows the results of two additional experiments. (B) Binding of FoxD3 to the Alb1 enhancer in ES cells. ChIP assays were performed with FoxD3 antibodies. Convergent arrows beneath the Alb1 map indicate PCR priming sites. Real-time PCR signals from control IgG ChIPs were subtracted, and the data were expressed as percent input values. Standard deviations from two replicate experiments are shown.