Abstract
Despite abundant examples of both adaptation at the level of phenotype and Darwinian selection at the level of genes, correlations between these two processes are notoriously difficult to identify. Positive Darwinian selection on genes is most easily discerned in cases of genetic conflict, when antagonistic evolutionary processes such as a Red Queen race drive the rate of nonsynonymous substitution above the neutral mutation rate. Genomic imprinting in mammals is thought to be the product of antagonistic evolution coincident with evolution of the placenta, but imprinted loci lack evidence of positive selection likely because of the ancient origin of viviparity in mammals. To determine whether genetic conflict is a general feature of adaptation to placental reproduction, we performed comparative evolutionary analyses of the insulin-like growth factor II (IGF2) gene in teleost fishes. Our analysis included several members of the order Cyprinodontiformes, in which livebearing and placentation have evolved several times independently. We found that IGF2 is subject to positive Darwinian selection coincident with the evolution of placentation in fishes, with particularly strong selection among lineages that have evolved placentation recently. Positive selection is also detected along ancient lineages of placental livebearing fishes, suggesting that selection on IGF2 function is ongoing in placental species. Our observations provide a rare example of natural selection acting in synchrony at the phenotypic and molecular level. These results also constitute the first direct evidence of parent–offspring conflict driving gene evolution.
Keywords: genomic imprinting, parent-offspring conflict, placentation, positive selection, sexual antagonism
The parent–offspring conflict theory posits that in organisms where there is parental inequality in the allocation of resources to the production of offspring, genetic antagonism may be a potent selective force shaping modes of reproduction and development (1–4). According to the kinship theory of genomic imprinting, parent-specific gene expression in placental mammals and seed-bearing plants is an outcome of this conflict (5). The strongest evidence of parent–offspring conflict associated with the evolution of matrotrophy (i.e., mother-feeding) is that the growth factor, insulin-like growth factor II (IGF2), and its antagonistic receptor, IGF2r, are oppositely imprinted in eutherian mammals and marsupials (6–8). Nevertheless, placentation and imprinting likely evolved in the common ancestor to Eutheria and Marsupialia >100 million years ago (9, 10); therefore, evidence that these two genes evolved under positive selection has been difficult to discern. Any evidence for antagonistic coevolution of genes that may have coincided with the evolution of placentas in mammals is likely to have faded into the background of neutral mutation accumulated since the Cretaceous.
The primary amino acid sequence of IGF2 has evolved under strong purifying selection among vertebrates, with 57% identity/68% conserved changes between human and the elasmobranch, Squalus acanthius, who last shared a common ancestor >400 million years ago. IGF2 expression during embryogenesis has been reported for many vertebrates, including a wide variety of teleost fishes. IGF2 is a potent stimulator of cell proliferation in all vertebrates. In mammals, IGF2 is a key promoter of both fetal and placental growth, but after birth, its expression is abolished or becomes highly tissue-restricted. In contrast, teleosts express IGF2 throughout development and into adulthood.
The neotropical fish family, Poeciliidae, is comprised of ≈200 species, all of which, with one exception, give live birth (11). Most poeciliids are lecithotrophic (i.e., yolk-feeding); eggs are vested before fertilization with enough nutrients to support embryonic development through to parturition (12). However, placenta-like structures that foster postfertilization maternal provisioning have evolved in several poeciliid lineages independently (13). Among several closely related species of poeciliids, there is highly developed placentation, intermediate development, or no placenta at all, offering the opportunity to examine transitional forms in a relatively brief evolutionary window. Within the genus Poeciliopsis, placentation in some species has been estimated by relaxed molecular clock analysis to have evolved as recently as 750,000 years ago (13). Maternal provisioning is characterized with the matrotrophy index (MI), measured as the ratio of the dry mass of a newborn fish to that of the fertilized egg.
In a previous study, we examined the allelic expression profile of IGF2 in two placental poeciliids, Heterandria formosa (MI = 30–40) and Poeciliopsis prolifica (MI = 5–10) and found that, unlike placental mammals, both species showed balanced biallelic expression of IGF2 throughout embryogenesis (14). The lack of a parent-of-origin effect on the transcriptional regulation of IGF2 suggested that: (i) parent–offspring intragenomic conflict does not operate in these fish despite their having placentas; (ii) IGF2 is not involved in development of the placenta in these fish and is thus immune to the selective influence of parent–offspring conflict; or (iii) if parent–offspring conflict mediated by IGF2 has occurred, it must be manifest in other ways. To test these possibilities, we examined the spatiotemporal expression profile of IGF2 in the poeciliid placenta and examined the IGF2 protein coding sequence for evidence of Darwinian selection in egg-laying and livebearing teleosts.
Results
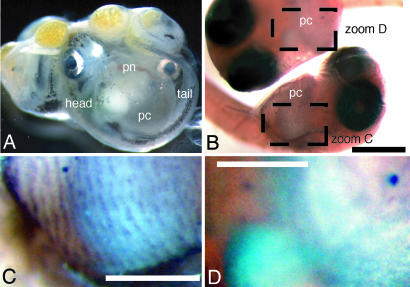
We performed RNA in situ hybridization to localize IGF2 expression in embryos of the livebearing placental poeciliid, H. formosa. Despite its independent origins in poeciliids, the placenta of matrotrophs is highly conserved in structure. The maternal component of the placenta is derived from the ovarian follicle, which becomes thickened and highly involuted. The fetal component of the placenta consists of a hypertrophied pericardial sac, comprising the highly vascularized portal network (15) (Fig. 1A, showing the matrotrophic poeciliid P. prolifica). As shown in Fig. 1 B–D, IGF2 transcripts are detected at high levels in interstitial cells of the portal network of the embryonic pericardium in H. formosa. These results confirm that the IGF2 gene is transcriptionally active in the poeciliid placenta, the most likely arena of parent–offspring intragenomic conflict, if such conflict exists in matrotrophic fishes.
Fig. 1.
RNA in situ hybridization detecting IGF2 transcripts in midgestation embryos. (A) Intact gravid ovary from P. prolifica female. (B) Ventral view of H. formosa embryos stained with sense (Upper) and antisense (Lower) probes. (Scale bar, 1 mm.) (C) Zoom C box of antisense from B showing staining of vascular interstices of the pericardial sac. (Scale bar, 0.5 mm.) (D) Zoom D box of sense from B. (Scale bar, 0.5 mm.) pn, portal network; pc, pericardium.
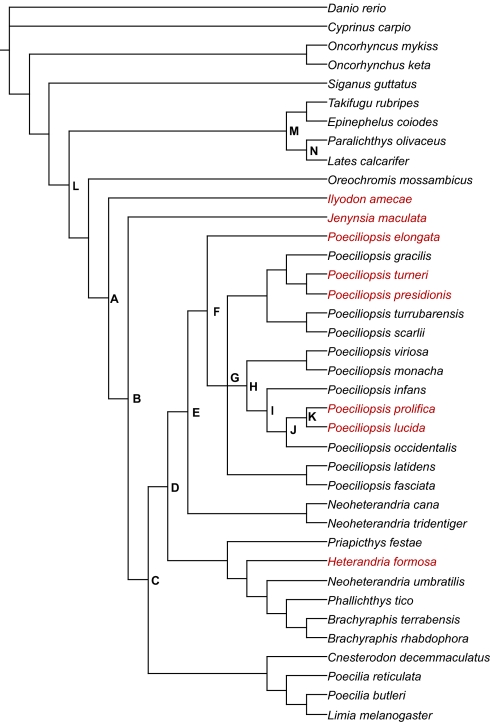
To examine more closely the potential effects of differing reproductive strategies on the evolution of IGF2, we assembled complete protein-coding sequences of this gene from 38 species of teleosts including 10 egg-laying species and 28 livebearing species (Fig. 2 and SI Table 4). We designed our analysis under the assumption that genetic antagonism influencing IGF2 evolution may be discernable within the overall pattern of evolution of the IGF2 gene sequence in teleosts. The three types of evolution that may operate on a gene sequence, neutral, purifying selection, or positive selection, can be discerned by statistical methods that compare the rate of nonsynonymous substitutions (amino acid replacing) per nonsynonymous site (dN) to the rate of synonymous (silent) substitutions per synonymous site (dS) (16). dN and dS are calculated from multiple sequence alignments of the gene of interest. In theory, a dN/dS ratio (represented by ω) will be equal to 1 if there is no constraint on the codon sequence, that is, the sequence is evolving neutrally. Purifying selection operating on a gene will result in ω<1, with strong purifying selection driving ω toward 0. Protein-coding genes, in general, evolve under strong purifying selection, because random nonsynonymous mutations will most likely diminish function rather than enhance it (17) (see SI Table 5). In the rare cases where nonsynonymous changes result in a fitness advantage, dN may exceed dS, resulting in ω>1 (positive selection). In practice, positive selection can be difficult to detect, because it may have operated only on specific codon sites within a gene, within specific lineages of a phylogeny, or within relatively brief and remote time periods.
Fig. 2.
Phylogenetic tree for all species used in the PAML analysis. Tree topology inferred from mitochondrial cytochrome b nucleotide sequences generated by Mr. Bayes. 3.1. Species in red represent those with extensive matrotrophy: MI >2. A-M node labels are presented in Table 2.
The phylogenetic analysis by maximum likelihood (PAML) software package uses maximum-likelihood statistics to test various evolutionary models for fit to a data set consisting of aligned sequence of a gene from a collection of species and a phylogenetic tree describing the history of divergence of the collection (18). Likelihood values (probability of the data given a particular model) are calculated with certain parameters fixed and others estimated from the data (free). The relative goodness of fit of a pair of given models, one that allows positive selection and one that does not, constitutes a test for the occurrence of positive selection on a gene. Models tested include those that account for selection on specific codon sites, within specific lineages, or the combination of selection on specific sites within specific lineages (see ref. 19 for full description of models). Because most mutations are either neutral or deleterious, models allowing only for neutral evolution (ω = 1) and purifying selection (0<ω<1) serve as the null hypothesis. Positive selection models allow an additional class of sites with ω>1. Positive selection models are compared with neutral/purifying models in a likelihood ratio test: 2 × the difference of the log likelihood values (2Δl) for the two models applied to a χ2 distribution to determine significance (P value). If a positive selection model provides a statistically significant better fit to the data, then the neutral/purifying model is rejected.
We first performed tests for selection on codon sites of IGF2 within all teleosts by comparing model M1a to M2a and M7 to M8 (19). The neutral/purifying model (M1a) allows only two site classes for ω: 0<ω0<1; and ω1 = 1. ω0 and the proportion of sites with ω0 (p0) are free parameters, whereas ω1 and the proportion of sites with ω1 (p1 = 1−p0) are fixed. The positive-selection model, M2a, allows a third class of sites with ω2>1, with ω2 estimated from the data. In this model ω0, ω2, p0 and p1 are free parameters, whereas ω1 and p2 (p2 = 1−p0−p1) are fixed. Models M1a and M2a average ω0 for all of the sites in this class. Sites under purifying selection, however, will likely vary a great deal with regard to how tolerant they are of nonsynonymous changes. For this reason, the model M7 was formulated to apply a continuous probability distribution (β) to the calculation of ω0. The β distribution can take many shapes in the interval 0–1 and therefore more accurately represents the distribution of ω0 values for this class. Model M8 is an extension of M7 that allows a third class of sites with ω2>1, with ω2 estimated from the data. Both model M2a and M8 apply “Bayes empirical Bayes” analysis (20) to the data set to determine the probability that a particular codon site falls within a particular class. In this way, sites under positive selection (i.e., ω2 class) can be identified.
Model M2a detects positive selection on one site across the teleost tree (site 139) with >95% probability (Table 1). However, this model calculates ω2 = 1 and generates a likelihood value identical to that of the neutral/purifying model M1a (P = 1). This test suggests, therefore, that one IGF2 codon may be evolving under positive selection in teleosts, but that the strength of selection, as measured by ω2, is very weak (averaged out to 1). In the comparison of models M7 and M8, the model allowing positive selection at specific sites fits the data significantly better than the neutral/purifying selection model (P = 0.0027). M8 estimates that 1.3% of sites (p2) fall in the positive selection class with ω2 = 2.63. This means that for the codons in this class, nonsynonymous mutations are more than twice as likely to become fixed than are silent mutations. M8 also identifies codon 139 as being under positive selection with >99% probability. This second test for selection, therefore, indicates that at least one IGF2 codon within teleosts has been subject to positive Darwinian selection with strong statistical support. Site-specific models, however, average ω2 across the whole phylogeny and cannot determine whether the positively selected sites have evolved under selection over the entirety of teleost history or have experienced a burst of strong selection along certain lineages.
Table 1.
Tests for selection on IGF2 in teleosts
| Model | Model comparison | Foreground branches | 2Δl | df | P value | ω* | Positively selected sites† |
|---|---|---|---|---|---|---|---|
| Site models | M1a vs. M2a | NA | 0 | 2 | 1 | 1 | 139 |
| M7 vs. M8 | NA | 11.82 | 2 | 0.0027 | 2.63 | 139 | |
| Branch-site models | Test 1/Test 2 | Poeciliidae | 28.27/3.26 | 2 | 7.26 × 10−7/0.0712 | 2.07 | 139, 146 |
| Test 1/Test 2 | Extensive matrotrophs | 24.26/7.81 | 2 | 5.40 × 10−6/0.0052 | 4.11 | 84, 86 | |
| Test 1/Test 2 | Extensive matrotrophs with P. lucida and P. prolifica | 21.85/8.64 | 2 | 1.80 × 10−5/0.0033 | 6.69 | 83, 86 |
Likelihood values are generated by using PAML 3.14b and are analyzed by a likelihood ratio test (2Δl) applied to a χ2 distribution with df = 1 − number of free parameters for each model. ″Extensive matrotrophs″ means that all species with extensive matrotophy (MI > 2), shown in red in Fig. 2, are designated as foreground lineages; ″P. prolifica and P. lucida″ means P. prolifica, P. lucida are included as foreground. NA, not applicable.
*ω values are presented as estimated from the data (ω2; see text). Values in bold are those significantly supported (P < 0.05).ω2 for branch/site models are shown for foreground branches as listed.
†Positively selected codon sites as determined by Bayes Empirical Bayes (BEB) analysis. Plain text indicates >95% confidence, and bold text indicates >99% confidence.
To distinguish these possibilities, we next performed tests for selection using a branch/site model (Model A) for positive selection at specific codon sites within specific lineages. Like model M2a, Model A allows a codon site class with ω2>1 but only along specified branches of the phylogeny (called foreground branches). The authors of PAML recommend two likelihood ratio tests for selection using branch/site models: Test 1 compares the likelihood of Model A to the neutral model M1a but cannot always distinguish positive selection from relaxed constraint; Test 2 is more stringent and compares Model A to Model A with ω2 fixed at 1 (21). We tested several different models that grouped various species together as the foreground (data not shown) but only two, all Poeciliidae as foreground or all extensive matrotrophs as foreground, had strong statistical support in at least one test (Table 1). Both Tests 1 and 2 support the conclusion that IGF2 has evolved under strong selection in matrotrophs (ω2 = 4.11). This means that nonsynonymous mutations are over four times more likely to become fixed than are silent mutations at some sites in these lineages. Furthermore, the branch/site model identifies two sites with >95% probability of evolving under positive selection in matrotrophs (Table 1 and Fig. 3). These two sites are immediately adjacent to the core binding motif of IGF2 for the type I IGF receptor. In addition to these two sites, high probabilities for positive selection (75–94.5%) were predicted for additional sites that clustered just downstream of the D/E domain boundary (Fig. 3B). In all vertebrates, the IGF2 prepropeptide undergoes two essential proteolytic cleavages: one to remove the signal peptide and a second to remove the E domain to produce the mature hormone (Fig. 3A) (22, 23).
Fig. 3.
Amino acid sites under positive selection in the IGF2 prohormone in matrotrophic fishes. (A) Schematic of domain structure of the IGF2 peptide sequence. (B) Schematic representation of a portion of IGF2 encompassing all amino acid sites exhibiting a >50% posterior probability (by Bayes Empirical Bayes analysis) of positive selection according to the branch-site models for both extensive matrotrophs (Table 1) and lecithotrophs (Table 3). Gray bars, sites with 50% < posterior probability < 95%; blue bars, sites with posterior probability > 95%: red bars, sites with posterior probability > 99%.
Two extensive matrotrophs, Poeciliopsis lucida and P. prolifica, were left out of the initial PAML analysis because of a large block of amino acid replacement encompassing fifteen contiguous codons. The replacement, in both, is the result of an insertion/deletion (indel) event at the 3′ end of exon 2, which encodes the proteolytic cleavage site separating the D and E domains (Fig. 4A). Examination of intronic sequence in several Poeciliopsis species reveals that the new codons were recruited from adjacent intron sequence and likely results from a single dramatic mutational event in their common ancestor. Because PAML cannot distinguish a single multicodon replacement from single-nucleotide changes, inclusion of P. lucida and P. prolifica IGF2 sequence gave a highly skewed estimation of ω in tests for selection. Nevertheless, the transition from lecithotrophy to matrotrophy in the common ancestral lineage of P. lucida and P. prolifica represents the most recent adaptation (≈750,000 years ago) to placental reproduction known in this family (13). We therefore repeated the branch/site PAML analysis including these two species but excluding the portion of IGF2 sequence encompassing the indel (Table 1). Again, the branch/site test for selection supports the conclusion that IGF2 codons near the receptor-binding motif are evolving under positive selection in matrotrophs. The >99% probability of positive selection on codon 86 is reinforced, but codons 83–87 all show >80% probability (Fig. 3B). Adding these two recently evolved matrotrophs to the data set also resulted in a dramatic increase in ω (ω2 = 6.69).
Fig. 4.
IGF2 E domain mutations in Poeciliopsis. (A) Sequence alignment of P. monacha, P. prolifica, and P. lucida illustrating the deletion of the 3′ 15 bases of exon 2 in P. prolifica and P. lucida. The deletion results in the recruitment of sequences that would have been intronic and utilization of a cryptic intronic splice donor site. (B) Conserved site of proteolysis to remove the E domain from the IGF2 prohormone. The nonconservative change (S→A) in P. infans is highlighted in yellow.
Although the tests for selection have confirmed that IGF2 codons have evolved under positive selection in matrotrophic fishes, they have not explicitly confirmed Darwinian selection driven by parent–offspring conflict. Except in instances of sustained selective pressure, positive directional selection is generally detectable only among recently diverged species when the rate of fixation of advantageous nonsynonymous changes briefly outpaces the neutral mutation rate. The positive selection on IGF2 in P. prolifica adheres to this burst-like pattern, (Table 2). A long period of purifying selection was interrupted by a burst of fixation of nonsynonymous changes coinciding with the evolution of placentation (branch J–K). In contrast, fixation of nonsynonymous changes in IGF2 has remained strong over a long evolutionary time frame in two ancient matrotrophic lineages in Cyprinodontiformes: Goodeinae, represented by I. ameca; and Jenynsiinae represented by J. maculata (Table 2 and Fig. 2). Both families comprise only viviparous species, with Goodeinae thought to have diverged from an ovoviviparous ancestor ≈16.5 million years ago (24). The surfeit of nonsynonymous changes along these two lineages suggests that positive selection on IGF2 did not occur as a burst followed by relative stasis but has continued unabated since the early adaptation to placental reproduction. Selection pressure is sustained, therefore, in lineages in which matrotrophy has evolved. By contrast, IGF2 has evolved under purifying selection in the purely egg-laying lineage leading to Lates calcarifer.
Table 2.
Substitutions along branches
| Node to node | n* | s† |
|---|---|---|
| A–I. ameca | 21.5 | 13.5 |
| A–J. maculata | 11.5 | 6.5 |
| A–P. prolifica | 11 | 13 |
| A–B | 1 | 2 |
| B–C | 4 | 4 |
| C–D | 1 | 2 |
| D–E | 0 | 1 |
| E–F | 0 | 0 |
| F–G | 0 | 1 |
| G–H | 0 | 1 |
| H–I | 0 | 0 |
| I–J | 0 | 1 |
| J–K‡ | 4(15)§ | 1 |
| K–P. prolifica | 1 | 0 |
| L–L. calcarifer¶ | 3 | 7 |
| L–M | 1 | 0 |
| M–N | 2 | 2 |
| N–L. calcarifer | 1 | 5 |
Substitutions compiled from reconstructed ancestral sequences generated by PAML for the teleost phylogeny in Fig. 2.
*n, nonsynonymous substitutions.
†s, synonymous substitutions.
‡Branch along which placentation emerged from ovoviviparous ancestor.
§() includes indel comprising 15 amino acid substitutions.
¶Egg-layer, example.
When all Poeciliidae were designated as foreground, branch/site Test 1 detected positive selection on two sites with ω = 2.07 (Table 1). We repeated the PAML analysis removing all matrotrophs to see whether positive selection operates on IGF2 in lecithotrophic species alone (Table 3). Both branch/site Tests 1 and 2 support a conclusion of positive Darwinian selection in lecithotrophic livebearers with ω = 2.27. Sites with >95% probability of being positively selected cluster near the proximal end of the E domain (Fig. 3B).
Table 3.
Tests for selection on IGF2 excluding matrotrophs
| Model | Model comparison | Foreground branches | 2Δl | df | P value | ω* | Positively selected sites† |
|---|---|---|---|---|---|---|---|
| Site models | M1a vs. M2a | NA | 0 | 2 | 1 | 1 | None |
| M7 vs. M8 | NA | 6.0 × 10−6 | 2 | 0.9997 | 1.00 | None | |
| Branch-site models | Test 1/Test 2 | Poeciliidae | 48.09/5.98 | 21 | 3.60 × 10−11/0.0203 | 2.27 | 122, 135, 139, 140, 145, 146, 147 |
Tests for selection on IGF2 in teleosts (excluding matrotrophs). Table 1 caption applies. NA, not applicable.
The PAML analysis shows positive selection of IGF2 in Poeciliidae to be concentrated on two regions of the peptide: (i) just proximal to the Type 1 receptor-binding site in matrotrophs and (ii) within the E domain just distal to the D/E proteolysis site in all livebearers. The latter suggests that Darwinian selection may be operating either on E peptide function or on processivity of the prohormone. The replacement of 15 contiguous codons of the E peptide by adjacent intronic sequence in the ancestor of P. lucida and P. prolifica suggests that E peptide function is not critical. Such a dramatic change in primary structure adjacent to the site of proteolysis, however, could ultimately affect the level of functional hormone by altering interaction of the endoprotease with its target site. Further support for selection on prohormone processing comes from a species closely related to these two (Fig. 2). Poeciliopsis infans exhibits a nonconservative amino acid substitution in the core of the prohormone cleavage site. This site is encompassed within a 5-aa motif that is invariant from zebrafish to human (Fig. 4B). Given the likelihood of compensatory evolution of other genes in these species, it would be difficult to ascertain the effect of these mutations on fetal growth in the fishes. In human fetuses, however, deficiency in posttranslational processing of the IGF2 prohormone and persistence of the large BCADE form leads to intrauterine growth retardation (23).
Discussion
In the mammalian placenta, the IGF2 peptide hormone promotes the proliferation and migration of fetal trophoblast cells as they invade the maternal decidua. IGF2 signaling, therefore, is at the crux of material exchange between mother and fetus (25, 26). Placental fishes lack a trophoblastic cell lineage per se, but the equivalent function is carried out by the hypertrophic highly vascularized embryonic pericardium. Our observation that IGF2 is highly expressed in the interstitium of the embryonic pericardium of H. formosa suggests this hormone is playing an analogous role in the growth and development of the poeciliid placenta. The observation that this gene has been subject to strong Darwinian selection in synchrony with the evolution of the placenta in matrotrophic teleosts underscores the central role of this gene in the regulation of vertebrate embryonic growth. When matrotrophs are removed from the evolutionary rate analysis, however, inflated ω values for IGF2 are still detected among lecithotrophic livebearers. The simple retention of fertilized eggs within the mother, even in the absence of direct maternal/fetal exchange, may afford the paternal genome the opportunity to impact maternal fitness by manipulating growth rates and gestational duration (27).
According to the kinship theory, the parent-specific gene expression characteristic of genomic imprinting is the result of an intragenomic arms race over maternal provisioning to offspring (5). The epigenetic nature of genomic imprints means they exist in a privileged arena for conflict: the erasure and sex-specific resetting ensures that offspring do not suffer from the selfishness of the parent of the opposite sex. Contrarily, a genetic mutation that alters IGF2 primary sequence, for instance creating a new allele advantageous to a father through enhancement of maternal provisioning, would seem to have an immediate negative affect on his daughters that inherit it. This would seem a powerful force against fixation of such an allele. However, Haig has shown this not to be the case in the formulation of his gestational drive hypothesis (28). Such an allele (D) can spread in a population if the fewer but fitter offspring of a Dd mother that inherit the D allele outcompete the more numerous but less-fit offspring of a dd mother.
The signal for positive selection on IGF2 in matrotrophic teleosts is exceptionally strong for a single-copy gene with a highly conserved developmental function. Hughes (16) has defined three types of positive Darwinian selection detectable at the molecular level: balancing selection, diversifying selection within gene families, and directional selection between species. The third category has been the most difficult to detect, because adaptive evolution of genes accompanying species divergence is generally episodic in nature; adaptive mutations quickly fade into the background of accumulating neutral mutations. Examples of adaptive evolution of genes have come primarily from studies where: (i) positive Darwinian selection is inferred because of the fixation of novel mutations to which adaptive function can be attributed (29, 30); or (ii) adaptation is inferred because Darwinian selection on a gene sequence is measurable (31–35). Positive Darwinian selection on IGF2 constitutes an example of enduring directional selection on a gene accompanying the evolution of a complex trait and evinces the Red Queen race (36) fostered by parent–offspring conflict in placental species.
Materials and Methods
RNA in Situ Hybridization.
Embryos were fixed in 4% paraformaldehyde. RT-PCR product encompassing the last coding exon of H. formosa IGF2 was cloned into the TOPO II (Invitrogen, Carlsbad, CA) plasmid. Sense and antisense probes by in vitro transcription from Sp6 and T7 promoters incorporating DIG (Roche, Basel, Switzerland). Hybridization was performed according to ref. 37.
PAML.
PAML analysis was performed with PAML 3.14b essentially according to ref. 18. Sequences were aligned by using Clustal X (38) and corrected by eye based on the amino acid sequence. The reference tree was inferred mitochondrial DNA tree from refs. 39 and 13 with the addition of other teleost mitochondrial cytochrome b sequences from GenBank using Mr. Bayes, version 3.1 (40, 41) using the GTR sequence evolution model. Four Markov chains, three heated and one cold, were run simultaneously with random starting trees. The program codeml in the PAML package was implemented to generate tests for positive selection, and each model was run several times. Tests for selection on the nuclear gene RAG1 were run in parallel to IGF2 as a control nonplacental gene (see SI Tables 5–7).
Supplementary Material
Acknowledgments
We thank A. Meyer and J. Travis for samples and L. Stransbaugh and C. Nelson for editing the manuscript. This work was supported by grants from the National Science Foundation (to M.J.O., R.J.O, D.N.R.).
Abbreviations
- IGF2
insulin-like growth factor II
- PAML
phylogenetic analysis by maximum likelihood
- MI
matrotrophy index
- dN
rate of nonsynonymous substitutions per nonsynonymous site
- dS
rate of synonymous substitutions per synonymous site.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. may be found in supporting information (SI) Table 4].
This article contains supporting information online at www.pnas.org/cgi/content/full/0705048104/DC1.
References
- 1.Trivers R. Am Zool. 1974;14:249–264. [Google Scholar]
- 2.Wallis M. J Mol Evol. 1996;43:93–100. doi: 10.1007/BF02337353. [DOI] [PubMed] [Google Scholar]
- 3.Xie S, Green J, Bixby JB, Szafranska B, DeMartini JC, Hecht S, Roberts RM. Proc Natl Acad Sci USA. 1997;94:12809–12816. doi: 10.1073/pnas.94.24.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constancia M, Kelsey G, Reik W. Nature. 2004;432:53–57. doi: 10.1038/432053a. [DOI] [PubMed] [Google Scholar]
- 5.Haig D. Annu Rev Ecol Syst. 2000;31:9–32. [Google Scholar]
- 6.DeChiara TM, Robertson EJ, Efstratiadis A. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 7.Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 8.Haig D, Westoby M. Am Nat. 1989;134:147–155. [Google Scholar]
- 9.Freyer C, Zeller U, Renfree MB. J Exp Zool. 2003;299:59–77. doi: 10.1002/jez.a.10291. [DOI] [PubMed] [Google Scholar]
- 10.Springer MS, Stanhope MJ, Madsen O, de Jong WW. Trends Ecol Evol. 2004;19:430–438. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Rosen DE, Bailey RM. Bull Am Mus Nat Hist. 1963;126:1–176. [Google Scholar]
- 12.Thibault RE, Schultz RJ. Evolution (Cambridge, UK) 1978;32:320–333. doi: 10.1111/j.1558-5646.1978.tb00648.x. [DOI] [PubMed] [Google Scholar]
- 13.Reznick DN, Mateos M, Springer MS. Science. 2002;298:1018–1020. doi: 10.1126/science.1076018. [DOI] [PubMed] [Google Scholar]
- 14.Lawton BR, Sevigny L, Obergfell C, Reznick D, O'Neill RJ, O'Neill MJ. Dev Genes Evol. 2005;215:207–212. doi: 10.1007/s00427-004-0463-8. [DOI] [PubMed] [Google Scholar]
- 15.Turner CL. J Morphol. 1940;67:59–87. [Google Scholar]
- 16.Hughes AL. Adaptive Evolution of Genes and Genomes. New York: Oxford Univ Press; 1999. [Google Scholar]
- 17.Kimura M. Proc Natl Acad Sci USA. 1991;88:5969–5973. doi: 10.1073/pnas.88.14.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z. Curr Opin Genet Dev. 2002;12:688–694. doi: 10.1016/s0959-437x(02)00348-9. [DOI] [PubMed] [Google Scholar]
- 19.Bielawski JP, Yang Z. Maximum Likelihood Methods for Detecting Adaptive Protein Evolution. New York: Springer; 2005. [PubMed] [Google Scholar]
- 20.Yang Z, Wong WS, Nielsen R. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Nielsen R, Yang Z. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 22.Duguay SJ. Horm Metab Res. 1999;31:43–49. doi: 10.1055/s-2007-978697. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Q, Basak A, Mbikay M, Tsang BK, Gruslin A. Proc Natl Acad Sci USA. 2005;102:11047–11052. doi: 10.1073/pnas.0502357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb SA, Graves JA, Macias-Garcia C, Magurran AE, Foighil DO, Ritchie MG. Mol Phylogenet Evol. 2004;30:527–544. doi: 10.1016/S1055-7903(03)00257-4. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton GS, Lysiak JJ, Han VK, Lala PK. Exp Cell Res. 1998;244:147–156. doi: 10.1006/excr.1998.4195. [DOI] [PubMed] [Google Scholar]
- 26.Constancia M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, Stewart F, Kelsey G, Fowden A, Sibley C, Reik W. Nature. 2002;417:945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 27.Crespi BJ, Semeniuk C. Am Nat. 2004;163:635–653. doi: 10.1086/382734. [DOI] [PubMed] [Google Scholar]
- 28.Haig D. Proc Natl Acad Sci USA. 1996;93:6547–6551. doi: 10.1073/pnas.93.13.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant PR, Grant BR. Science. 2006;313:224–226. doi: 10.1126/science.1128374. [DOI] [PubMed] [Google Scholar]
- 30.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- 31.Hughes AL. Genetics. 1991;127:345–353. doi: 10.1093/genetics/127.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metz EC, Palumbi SR. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 33.Messier W, Stewart CB. Nature. 1997;385:151–154. doi: 10.1038/385151a0. [DOI] [PubMed] [Google Scholar]
- 34.Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. Proc Natl Acad Sci USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurle B, Swanson W, Green ED. Genome Res. 2007;17:276–286. doi: 10.1101/gr.6004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Valen L. Evol Theor. 1973;1:1–30. [Google Scholar]
- 37.Xu QA, Wilkinson DG. In: In Situ Hybridization: A Practical Approach. Wilkinson DG, editor. New York: Oxford Univ Press; 1998. pp. 87–106. [Google Scholar]
- 38.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 39.Mateos M, Sanjur OI, Vrijenhoek RC. Evol Int J Org Evolution. 2002;56:972–984. doi: 10.1111/j.0014-3820.2002.tb01409.x. [DOI] [PubMed] [Google Scholar]
- 40.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 41.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.