Fig. 1.
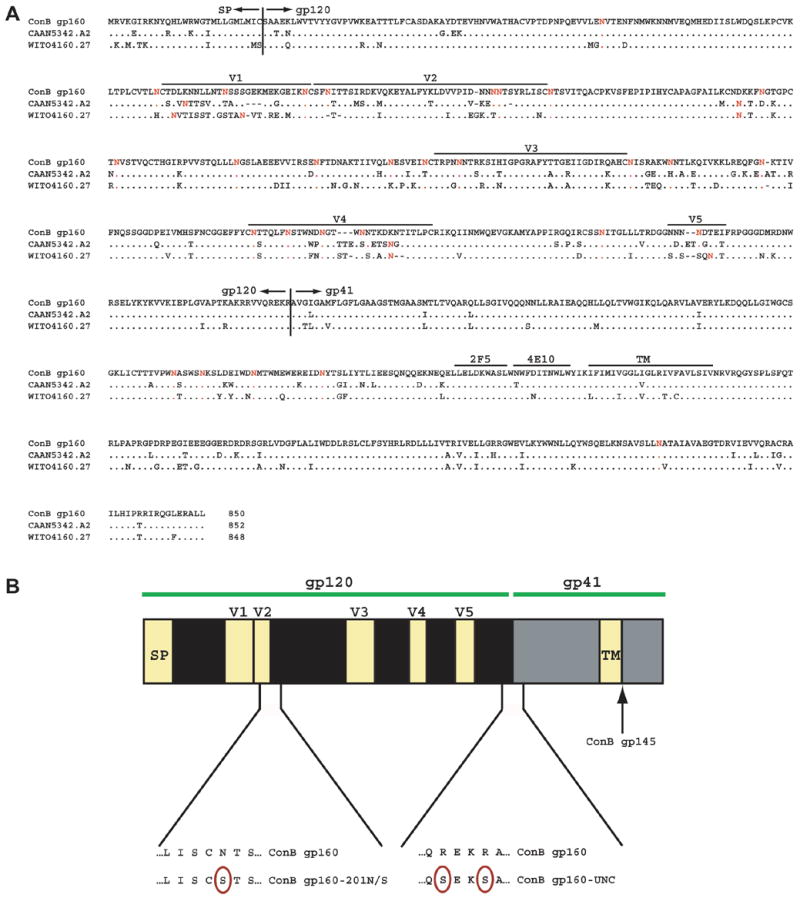
Generation of consensus subtype B (ConB) envelopes. (A) Alignment of the deduced protein sequence of full length (unmodified) ConB Env with those of recently transmitted, contemporary subtype B Env controls (CAAN5342.A2, WITO4160.27). Sequences are compared to ConB gp160, with dots indicating sequence identity, and dashes indicating gaps introduced for optimal alignment. Potential N-linked glycosylation sites are highlighted in red. The locations of signal peptide (SP) and gp120/gp41 cleavage sites are indicated, as are the positions of the variable loops (V1–V5) and the transmembrane (TM) domain. (B) Location of specific mutations within the ConB Env sequence. Amino acid substitutions are highlighted (see text for design and construction details of ConB gp160-201N/S, ConB gp160-UNC and ConB gp145).
