Figure 7.
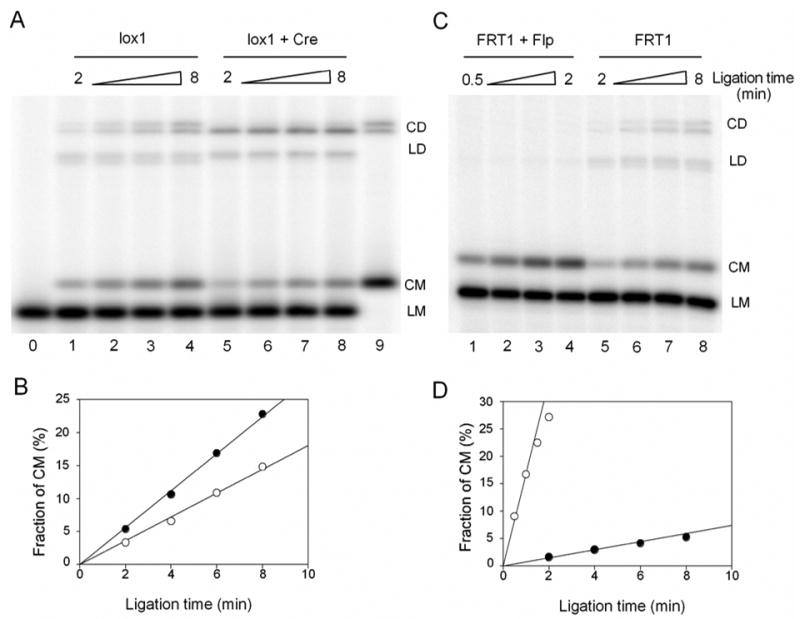
Cyclization experiments of 200bp gapped DNA and their complexes bound with Cre or Flp recombinases. (A) 1nM lox1 in the absence of Cre (lanes 1–4) and in the presence of 20nM Cre (lanes 5–8) was ligated simultaneously (but in separate reactions) by the same amount of T4 DNA ligase. Aliquots of the ligation mixture were quenched after 2, 4, 6, 8 minutes of the reaction. Lane 0 and 9 are unligated and completely ligated lox1 samples. CD, circular dimer; LD, linear dimer; CM, circular monomer; LM, linear monomer. (B) The accumulations of CM for lox1 DNA with (○) or without (●) bound Cre. Fraction of CM was calculated as the ratio of CM to the DNA substrate for each lane from the phosphorimages. For the experiment shown here, the cyclization rate of the Cre-bound DNA (○) was 64% of that of the protein-free fragment (●). (C) 1nM FRT1 in the presence (lanes 1–4) and in the absence of 20nM Flp (lanes 5–8) was ligated simultaneously (but in separate reactions) by T4 DNA ligase. The ligase concentration during the ligation of the Flp-bound fragment was 2.5 times lower than that for the protein-free fragment, to produce proper amounts of CM for quantitation. Also note that the time course in lanes 1–4 was 4 times shorter than in lanes 5–8. (D) The accumulations of CM for FRT1 DNA with (○) or without (●) bound Flp. The fraction of CM in the second plot (●) has been normalized to compensate for the 2.5 fold difference in the actual ligase concentrations. For the experiment shown here, the rate of cyclization of the Flp-bound fragment was 22 times higher than the corresponding rate for protein-free fragment.
