Figure 2. Origin of DNA duplexes used in this binding study.
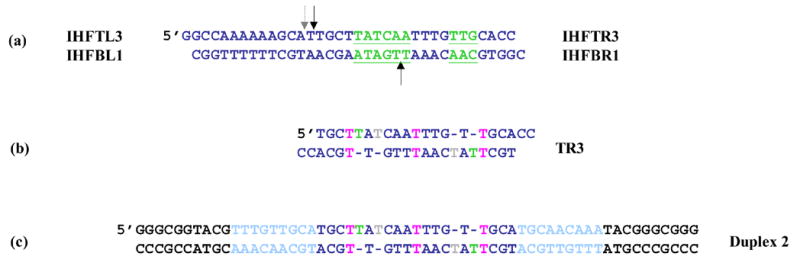
(a) IHF’s cognate site, H’. The IHF consensus site is underlined in green. The black arrows show the location of backbone nicks placed 9 base pairs apart in an effort to phase AHU binding for homogeneous crystal formation. The dashed arrow shows the location of the nick in the IHF-DNA crystal structure (1IHF). (b) DNA observed in an AHU-DNA cocrystal (1P71). Two copies of the top right oligonucleotide bound to each other forming a duplex with 4 unpaired Ts and 3 T • T mismatches. Color coding is as follows: green, analogous base in crystal structure is stacked; grey, analogous base in crystal structure is flipped; pink, T • T base pair (c) DNA duplex for gel shift experiments. The middle of the site is identical to duplex TR3 from the crystal. The flanking DNA in light blue is identical to DNA from neighboring protein-DNA complexes that contact AHU in the crystal. The black flanking sequence destabilizes a hairpin that forms in its absence and complicates binding experiments.
