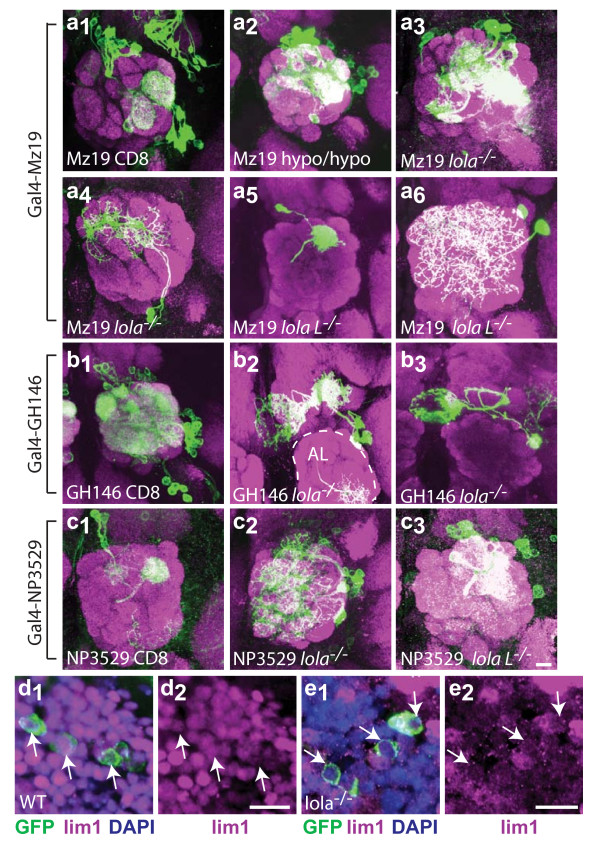
Figure 7.
Evidence that lola regulates expression of Gal4 drivers and Lim1. (a) Gal4-Mz19 mislabeling analysis. Whole-animal Gal4-Mz19, UAS-GFP::CD8 expression labels a small subset of PNs (a1). See Figure 2 for Mz19 and GH146 MARCM controls. Mutations in lola result in the labeling of more cell bodies than Mz19 normally labels (hypo: ore5D2) (a2, a3), as well as the labeling of ectopic cell types such as local interneurons (LNs) (a4, a6) and DL1 that is normally labeled by Gal4-GH146 but not Gal4-Mz19 (a5). (b) lola-/- Gal4-GH146 clones frequently mislabel a set of neurons in the region of the AL that project to the medial lobe of the MB and to the central body complex. (c) NP3529 mislabeling analysis. NP3529 labels two dorsal PNs that project to DL1 (c1). lola-/- clones dramatically increase the number of cells labeled near the AL, including cells in the lateral lineage and LNs (c2). lolaL-/- clones show similar mislabeling (c3). anti-CD8::GFP in green, anti-nc82 neuropil in magenta. (d, e) lola regulates Lim1 expression in vPNs. Wild-type cells in the region of the antennal lobe, including most (at least four) vPNs express Lim1 (d). In lola-/- cells, Lim1 expression is lost in vPNs (e). Note that the lola-/-clone includes neighboring cells in addition to those labeled by Gal4-GH146, and many cells have lost Lim1 staining. White arrows mark the cell bodies of GFP positive cells. anti-CD8::GFP in green, anti-Lim1 in magenta, DAPI in blue. Scale bar, 10 μm.