TABLE 2.
Thermal unfolding parameters
| Variant | pH °C |
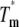 kcal mol−1 kcal mol−1
|
ΔHTm kcal mol−1 K−1 | ΔCP °C | Tx† kcal mol−1 | ΔGTx‡ |
|---|---|---|---|---|---|---|
| Wild-type | 6.0 | 68.9 ± 0.1¶ | 141 ± 2 | 3.9 ± 0.1 | 34.6 ± 0.1 | 7.2± 0.1 |
| 6.5 | 67.9 ± 0.1 | 147 ± 2 | 32.3 ± 0.1 | 7.8 ± 0.3 | ||
| 7.0 | 67.0 ± 0.2 | 140 ± 3 | 32.9 ± 0.1 | 7.2 ± 0.1 | ||
| 7.5 | 66.5 ± 0.1 | 163 ± 3 | 27.2 ± 0.1 | 9.6 ± 0.1 | ||
| 8.0 | 65.1 ± 0.2 | 133 ± 3 | 32.7 ± 0.1 | 6.5 ± 0.2 | ||
| S-126C | 6.0 | 69.3 ± 0.1 | 157 ± 9 | 4.0 ± 0.1 | 32.2 ± 0.1 | 8.7 ± 0.9 |
| 6.5 | 69.2 ± 0.1 | 144 ± 4 | 35.1 ± 0.1 | 7.3 ± 0.5 | ||
| 7.0 | 68.7 ± 0.1 | 148 ± 5 | 33.4 ± 0.1 | 7.8 ± 0.3 | ||
| 7.5 | 67.0 ± 0.1 | 139 ± 4 | 33.9 ± 0.1 | 6.9 ± 0.2 | ||
| 8.0 | 66.4 ± 0.2 | 137 ± 2 | 33.7 ± 0.1 | 6.7 ± 0.3 | ||
| S-265C | 6.0 | 63.1 ± 0.1 | 112 ± 5 | 3.3 ± 0.1 | 36.1 ± 0.1 | 4.6 ± 0.4 |
| 6.5 | 62.7 ± 0.1 | 110 ± 4 | 36.4 ± 0.1 | 4.4 ± 0.2 | ||
| 7.0 | 61.5 ± 0.2 | 109 ± 4 | 35.3 ± 0.1 | 4.3 ± 0.1 | ||
| 7.5 | 60.1 ± 0.1 | 108 ± 3 | 34.1 ± 0.1 | 4.2 ± 0.1 | ||
| 8.0 | 59.1 ± 0.1 | 104 ± 3 | 33.9 ± 0.1 | 4.0 ± 0.3 | ||
| S-126/265C | 6.0 | 64.6 ± 0.1 | 127 ± 7 | 2.9 ± 0.1 | 23.4 ± 0.1 | 7.9 ± 0.2 |
| 6.5 | 63.8 ± 0.1 | 112 ± 5 | 26.0 ± 0.1 | 6.7 ± 0.1 | ||
| 7.0 | 62.7 ± 0.3 | 101 ± 6 | 29.5 ± 0.1 | 5.1 ± 0.2 | ||
| 7.5 | 62.1 ± 0.1 | 95.6 ± 1 | 30.7 ± 0.1 | 4.6 ± 0.3 | ||
| 8.0 | 60.3 ± 0.3 | 93 ± 3 | 29.8 ± 0.1 | 4.3 ± 0.1 |
