Figure 2.
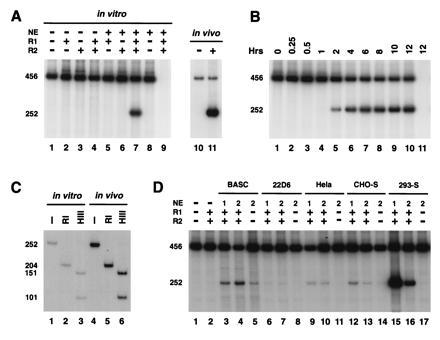
In vitro signal joint formation by deletion. (A) In vitro signal joint formation by deletion requires RAG1, RAG2, and nuclear extracts. pJH200 was incubated at 30°C with the indicated combinations of purified RAG1 (R1), RAG2 (R2), and nuclear extracts (NE) from 293-S cells under the conditions describe. The DNA was then purified and analyzed by PCR assay as described in Fig. 1. The autoradiograph from one such experiment is presented on the left (lanes 1–9), while the right (lanes 10 and 11) shows the 32P-labeled PCR products amplified from in vivo-recombined DNA purified from 293T cells transfected with pJH200 alone (−) or cotransfected with RAG1 and RAG2 (+). Lane 9 correspond to a control reaction, RAG1, RAG2, and NE but not template DNA. (B) Time course of in vitro signal joint formation. The reaction conditions were identical to those described in A, lane 7 (with RAGs and nuclear extract) and the products were analyzed as explained in Fig. 1. Lane 11 is identical to lane 10 except that template DNA was not included in the reaction. (C) Digestion pattern of the in vivo- and in vitro-recombined products. Purified 252-bp 32P-labeled PCR products were incubated with no enzyme (−), RsaI (RI), or HindIII (HIII) and separated on an 8% polyacrylamide/1× TBE gel. The 252-bp fragment contains a unique HindIII site 151 bp from the R14 oligonucleotide. This fragment also contains two RsaI sites, one in the 23 RSS and the other 25 bp from oligonucleotide R14. (D) Different nuclear extracts can complement RAG proteins in signal joint formation. RAG1 (R1) and RAG2 (R2) were supplemented with extracts prepared from BASC6C2 (BASC), 22D6, HeLa, CHO-S, or 293-S cells, as indicated. Each sample was processed as in A and the bands were visualized by autoradiography.
