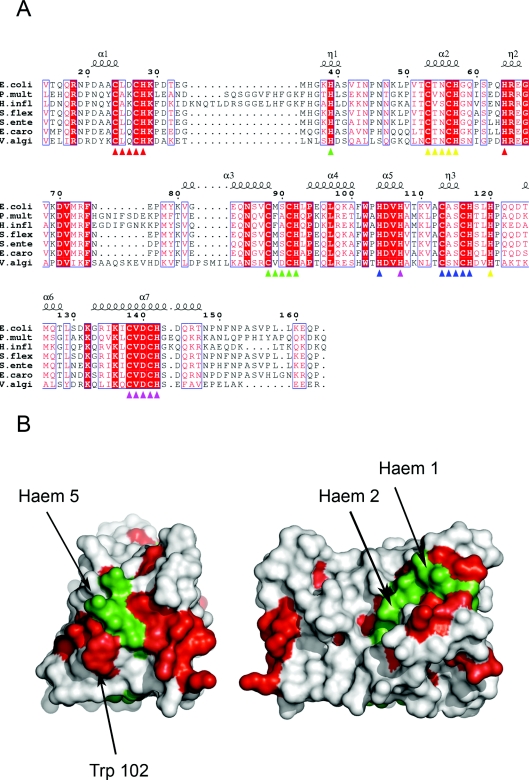
Figure 2. Amino acid sequence conservation in NrfB primary and tertiary structures.
(A) An amino acid sequence alignment of E. coli NrfB (residues 14–163) with NrfBs from Pasteurella multocida (P.mult), Haemophilus influenzae (H.infl), Shigella flexneri (S.flex), Salmonella enterica (S.ente), Erwinia carotovora (E.caro) and Vibrio alginolyticus (V.algi). Red-filled boxed amino acids are identical in the alignment, unfilled boxed amino acids are similar in the alignment. The secondary structure of E. coli NrfB is indicated above the sequence alignment. The CXXCH motifs are indicated by coloured triangles under the alignment according to: haem 1, red; haem 2, yellow; haem 3, green; haem 4, blue; haem 5, magenta. The corresponding distal histidine residues of each haem are also coloured accordingly. (B) Surface map of the side (right-hand panel) and C-terminal end (left-hand panel) of NrfB. Completely conserved residues are shown in red, haems 5, 2 and 1 are shown in green. All other residues are shown in white.