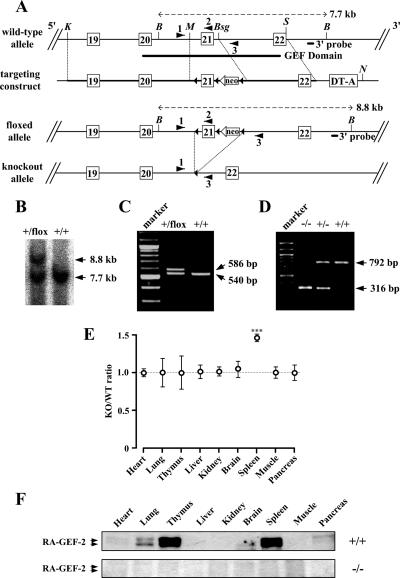
Figure 5.
Targeted disruption of the mouse RA-GEF-2 gene. (A) Schematic representation of the wild-type allele (RA-GEF-2+), the floxed allele (RA-GEF-2flox), and the knockout allele (RA-GEF-2−). Exons 19–22 (white rectangles), loxP sites (black triangles), TK-neo (neo), and diphtheria toxin A chain cassette (DT-A) are indicated in the targeting construct. The 3′ probe for Southern blot analysis, a trio of primers for genotyping by PCR, and restriction enzyme sites (K, KpnI; B, Bsu36I; M, MfeI; Bsg, BsgI; S, StyI; N, NotI) are also shown. (B) Southern blot analysis of the RA-GEF-2 gene. Genomic DNA was prepared from mouse tails, digested by Bsu36I, and hybridized with the 3′ external probe. The RA-GEF-2+ allele generated a 7.7-kb band, and the RA-GEF-2flox allele generated an 8.8-kb band. (C) PCR analysis of the leftmost loxP. Genomic DNA was prepared from mouse tails and analyzed by PCR. Primers 1 and 2 generated a 540-base pair band from RA-GEF-2+ allele and a 586-base pair band from the RA-GEF-2flox allele. (D) PCR-based genotyping of RA-GEF-2 knockout. Primers 1 and 3 generated a 792-base pair band from the RA-GEF-2+ allele and a 316-base pair band from the RA-GEF-2− allele. (E) Comparison of tissue-to-body weight ratios between RA-GEF-2−/− (KO) and wild-type (WT) mice. At least seven mice of each genotype were examined at the ages of 8 wk. ***p < 0.001. (F) Immunoblot analysis for RA-GEF-2. Protein extracts were prepared from tissues and used for immunoblotting by anti-RA-GEF-2 antibody.