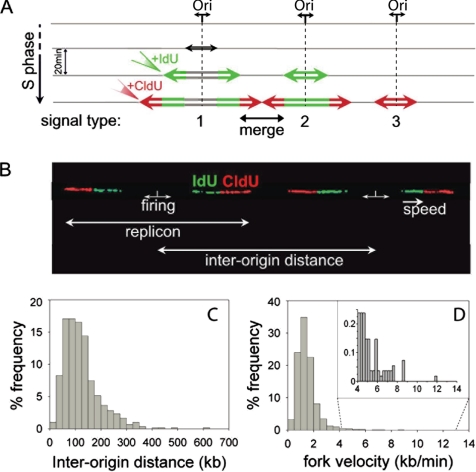
Figure 1.
Interorigin distances and total fork velocities. (A) Schematic representation of signals deriving from equal pulse-labeling with iodo-deoxyuridine (IdU) and chloro-deoxyuridine (CldU). Replication forks progress bidirectionally at the same rate from the origin and incorporate the analogues forming a symmetrical replication bubble. On detection of IdU and CldU, three types of signals may be obtained: a green and red signal with a gap between the green segments, corresponding to initiations that occurred before the beginning of the pulse (1); a dual-color signal with a continuous green segment corresponding to origins that fired during the first pulse (2); and a single isolated red signal corresponding to origins that fired during the second pulse (3). A continuous red signal flanked by two green ones is formed by the merging of two forks from adjacent origins. (B) Two adjacent replication bubbles are visualized on a combed DNA molecule (type 1 signal). The replication origins are assumed to be located in the midpoints of the unlabeled segments (see text). The distance between adjacent replication origins represents the interorigin distance. Fork speed is calculated by dividing the length of each fluorescent signal by the time of the pulse. (C) Histogram of the interorigin distances (median = 111; N = 606). (D) Histogram of the total fork velocities (mean = 1.46; N = 5460) for the human primary normal keratinocytes. In the panel, an enlarged view of the higher fork velocities (4–12 kb/min) is shown.