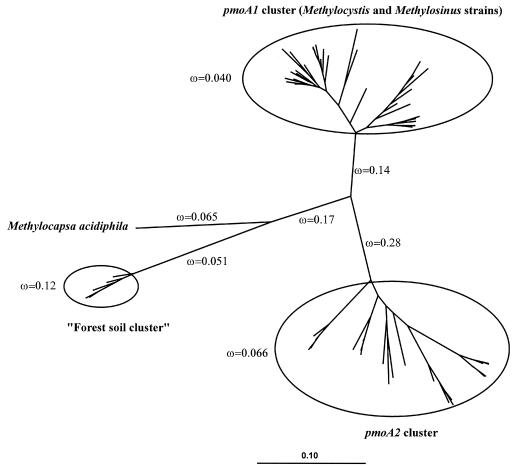
FIG. 5.
Analysis of ω values within an unrooted phylogenetic tree incorporating 61 selected pmoA nucleotide sequences. The branching topology was calculated by using a neighbor-joining algorithm (29) with a Jukes-Cantor correction (20) (2,000 data resamplings), and branch lengths were adjusted by the paml program. A maximum-likelihood algorithm produced nearly the same tree topology. The distance bar indicates 0.1 nucleotide substitution per position. The paml model (model 2 in codeml) was fit with eight separate ω values as indicated: one for each of the five longest branches and one each for the pmoA1 cluster, the pmoA2 cluster, and the “forest soil cluster,” respectively, which are enclosed by circles. The pmoA1 cluster included sequences from various Methylocystis and Methylosinus strains (accession no. U31569, U31651, AJ431386, AJ431388, AJ458994, AJ458998, AJ459000-AJ459003, AJ459008, AJ459011, AJ459013, AJ459014, AJ459019, AJ459021-AJ459025, AJ459027-AJ459029, AJ459034, AJ459036, AJ459038, AJ459046-AJ459048, and AJ459052). The pmoA2 cluster included sequences from pure cultures of Methylocystis and Methylosinus and sequences retrieved from soils by cultivation-independent methods (accession no. AJ431389, AJ299961, AY080950, and AJ459013 and sequences from the present study). The “forest soil cluster” contains sequences retrieved by Holmes et al. (18) using cultivation-independent methods (accession no. AF148525, AF148521, AF148527, AF148528, and AF148523).