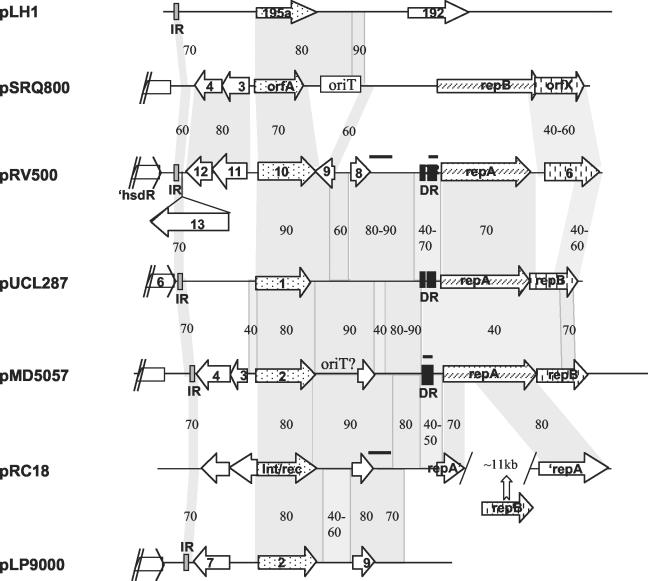
FIG. 4.
Sequence conservation among plasmids from LAB. Lfasta and Lalnview were used to detect and visualize nucleotide identity between plasmid sequences as schematized here (http://pbil.univ-lyon1.fr/lfasta.php). Gray shading and figures indicate sequence conservation and the percentage of nucleotide identity between two DNA segments. Segments with 100% nucleotide identity between pRV500 and pMD5057 or pRC18 are indicated by horizontal thick lines above the sequence lines. For easier visualization of sequence conservation, the DNA segment corresponding to orf13 of pRV500, which shares no similarity with other plasmids, was positioned underneath. Black and gray boxes represent stretches of direct repeats (DR) and palindromic sequences (IR), respectively. These palindromic sequences (about 40 bp) have higher conservation (>90% nucleotide identity) than that indicated according to the Lfasta program for the slightly larger region. orf genes putatively coding for recombinases and initiator proteins are indicated with dotted and hatched arrows, respectively, while those sharing amino acid similarity with orf6 of pRV500 are represented with arrows with vertical bars. Relevant portions of plasmids are schematized at the same scale here; for pLH1, pMD5057, and pLP9000, they are in the reverse direction relative to the entry in the database. Accession numbers for plasmid sequences (complete) were as follows: pLH1, AJ222725; pSRQ800, U35629; pUCL287, X75607; pMD5057, AF440277; pRC18, AF200347; pLP9000, AY096005.