FIG. 1.
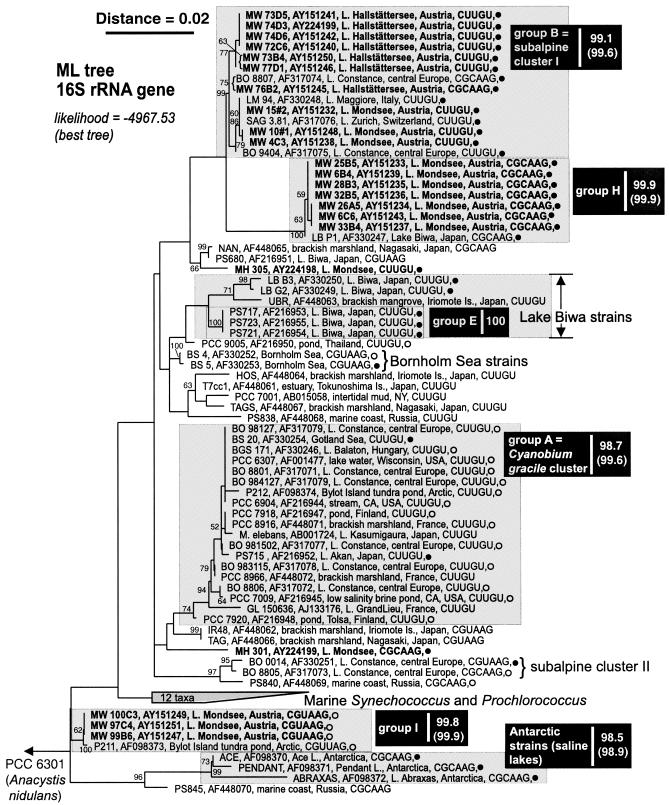
Maximum-likelihood tree of the picophytoplankton clade sensu Urbach et al. (38), inferred from 16S rRNA gene sequences (1,383 nt positions). Terminal branches display isolate and GenBank accession numbers (given in bold font for sequences determined in this study), isolation details (location, habitat), helix 49 tetraloop motif, and pigment group (•, PE rich; ○, PC rich). Numbers at nodes indicate the percent bootstrap frequency (1,000 replicates) obtained from neighbor-joining (31) trees calculated by TREECON for Windows (39) assuming a Jukes-Cantor model of nucleotide substitution. Bootstrap values of <50% are not shown. Minimum and mean (parentheses) pairwise percent similarities are shown on the right-hand side of each group label. Sources: results herein, references 10, 28, 29, and 40; Warwick Vincent, personal communication (isolation details for P211 and P212); and Watanabe, personal communication (isolation details for NAN, UBR, HOS, T7cc1, TAG, TAGS, IR48 PS-838, PS-840, and PS-845). The Ernst et al. (10) cluster designations subalpine cluster I, Lake Biwa strains, Bornholm Sea strains, Cyanobium gracile cluster, and subalpine cluster II are also provided. The outgroup was Synechococcus PCC 6301 (formerly Anacystis nidulans).
