Figure 2. KIS phosphorylates SF1 in vitro on serines 80 and 82.
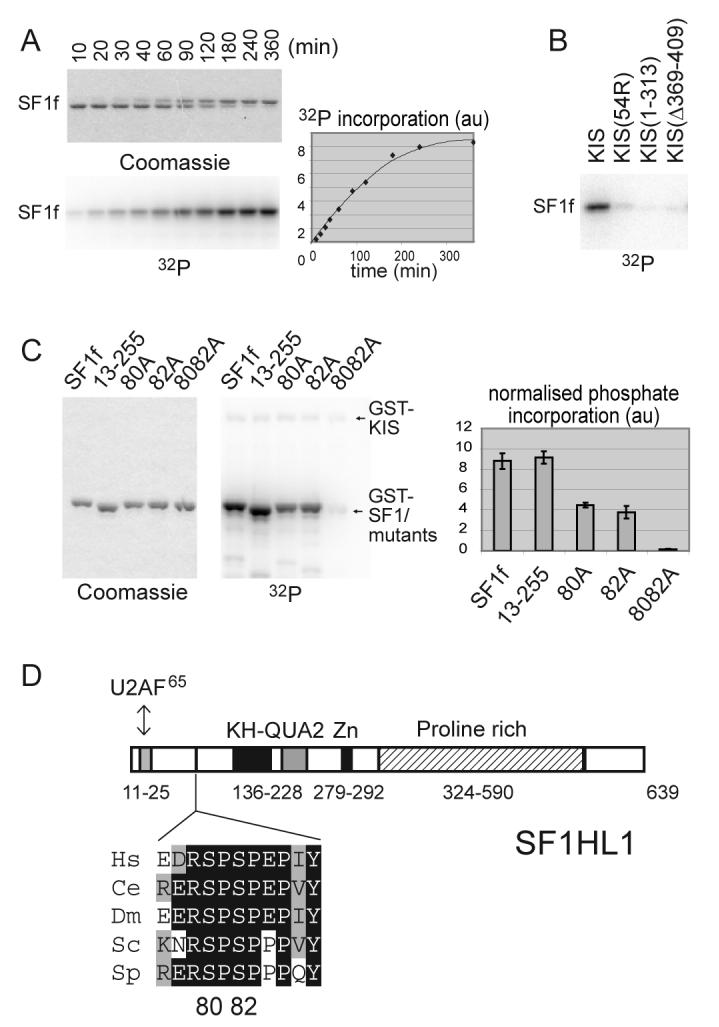
A) SF1[1-255] (SF1f) was phosphorylated by KIS with 10 μM [γ-32P]ATP for the indicated times. Reaction products were analysed by SDS PAGE, Coomassie blue staining and phosphorimaging. Phosphorylation of SF1f induced a shift of the SF1f band on SDS-PAGE.
B) The ability of similar amounts of the indicated forms of KIS to phosphorylate SF1f in vitro was compared, showing that deletion of or within the UHM domain of KIS impaired SF1f phosphorylation.
C) Phosphorylation in vitro of different forms of GST-SF1f as indicated. Reactions were stopped while phosphate incorporation was still linear with time (See “Experimental Procedures”). Mutation of both serines 80 and 82 completely abolished phosphate incorporation. Quantification: mean values of three experiments.
D) Primary structure of the SF1 isoform SF1HL1 (Alternatively spliced forms of SF1 differs by the length and sequence of their proline rich C-terminal region [28,51]). The SPSP motif is located in between the N-terminal U2AF65 binding region and the KH-QUA2 domain that mediates recognition of the BPS [20,52]. The SPSP motif is in a highly phylogenetically conserved region as presented in the alignment [15]. The sequences of SF1 are from the following organisms Hs, Homo sapiens; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.
