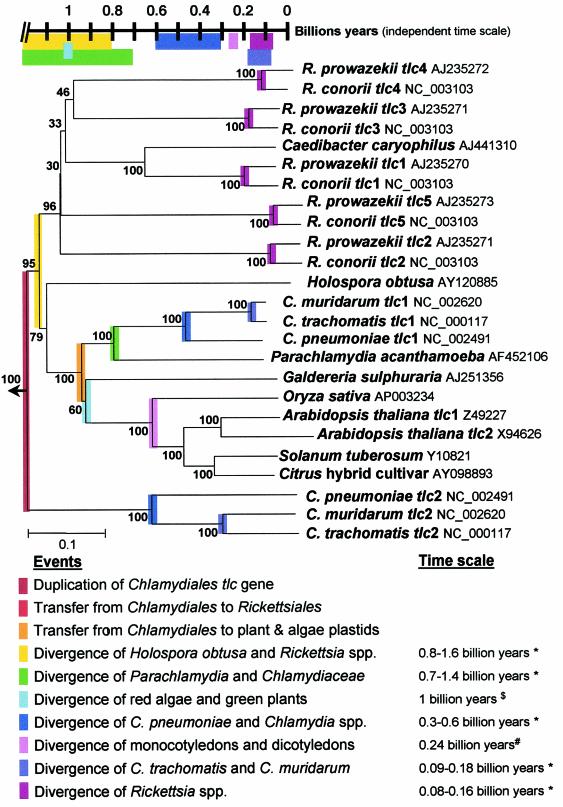
FIG. 1.
p-Distance neighbor-joining tree inferred from amino acid sequences of the ADP/ATP translocase of P. acanthamoebae, Chlamydiaceae, Rickettsiales, and plant and alga plastids. Bootstrap values resulting of 100 replications are present at branch points. The tree was rooted with four proteins present in E. cuniculi, whose functions are unknown and were identified by BLAST of nonmitochondrial ADP/ATP translocase. The time scale was derived from estimates obtained from the literature (13, 26) and on the basis of 16S rRNA gene sequence divergence (20). Note the congruence between the node of a given divergence (see neighbor-joining tree) and its estimated time (see time scale). ✽, Estimated from 16S rRNA divergence (20); $, estimated by Sogin et al. (26); #, estimated by Gale et al. (13).