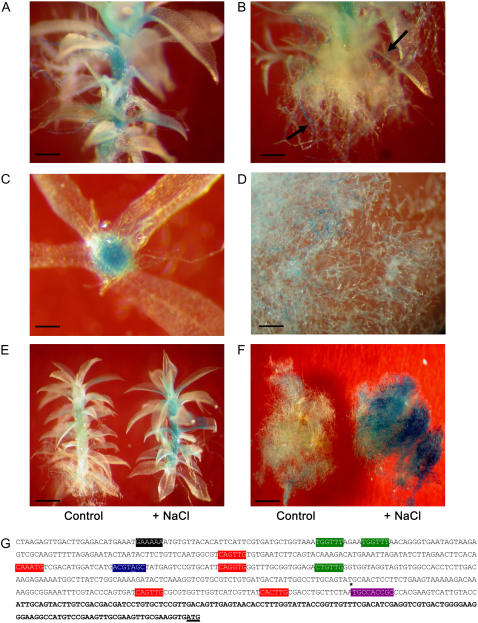
Figure 2.
Tissue-specific expression of PpENA1. The PpENA1 promoter (1,451 bp) was fused to a GUS-reporter gene and the construct transformed into Physcomitrella. A, GUS staining of a representative 5-week-old gametophyte grown on standard medium without NaCl, showing GUS staining in the stem and the basal part of the leaves. Bar = 0.4 mm. B, GUS staining of rhizoids from a nonstressed gametophyte. Arrows indicate the strongly stained filaments. Bar = 0.4 mm. C, Cross section of a gametophyte showing staining in the hydroids, cortex, and epidermis. Bar = 0.2 mm. D, Ubiquitous GUS staining in nonstressed protonemata. Bar = 0.4 mm. E, Comparison of GUS staining in two gametophytes originating from the same colony either left untreated or exposed to 100 mm NaCl for 24 h before GUS staining. The staining is significantly stronger, but still confined to the same tissues after salt stress. Bar = 0.6 mm. F, The effect of salt treatment on protonemata. A 10-d-old protonematal colony was divided in two and one-half of the colony was exposed to 100 mm NaCl for 24 h and the other half not treated. The strong GUS staining throughout the tissue is seen after NaCl stress. Bar = 1.5 mm. G, In silico PpENA1-promoter analysis. Putative cis-regulatory elements in the PpENA1 promoter were identified searching PLACE or NSITE-PL. Because the importance of regulatory elements generally decreases with increasing distance from the transcription start site, only the first 500 bp upstream of the putative transcription start site were included in the analysis. Putative GT-1 box (GAAAAA, black), Myb (WAACCA, CTGTTG, green), Myc (CANNTG, red), ABRE (ACGTGGC, blue), and CE-1 coupling elements (TGCCACCGC*, purple) are indicated. Mismatches in the consensus sequence are underlined and elements with an expected mean frequency <0.05 in 500 bp of random DNA sequence are marked with an asterisk. The putative 5′ UTR based on available EST sequences is shown in bold.