Figure 1.
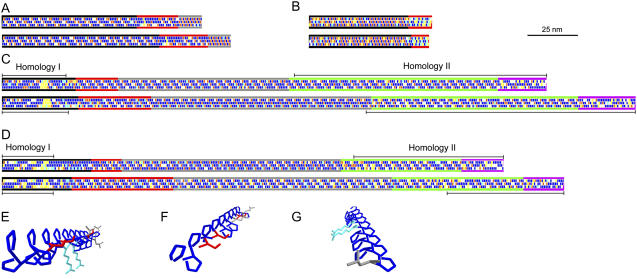
Diagrammatic representations of chimeric HRGP shaft domains from Chlamydomonas. A, Gp1. B, Vsp3. C, Sag1. D, Sad1 longitudinal face diagrams. Amino acids are depicted as rectangles along the three faces of PII helices (Ferris et al., 2005). Blue, Pro; red, Ser; light blue, charged; white, other. Yellow denotes guest amino acids: Two or more sequential amino acids other than Pro that have the potential to disrupt the helical configuration (Creamer, 1998). Diagrams at the top derive from C. reinhardtii, at the bottom from C. incerta. Subdomains are indicated by colored longitudinal lines as follows: A, Black, main shaft; red, kink; gray, neck. B, Black, main shaft; red, P3X3 subdomain. C and D, Black, 2A; red, 2B; gray, 2C; green, 2D; purple, 2E (Ferris et al., 2005). Homology I and II regions are described in text. E to G, Three-dimensional reconstruction of a PII helix from the PPPPPSPPSPRPPRPPPLPPSPPPPLL sequence of the 2A subdomain of Sag1 C. reinhardtii, emphasizing the three longitudinal faces. The γ-carbon-to-γ-carbon spacing of Pro amino acid residues along a PII longitudinal face is calculated to be 0.934 nm; Pro spacing along a PII helical face is calculated to be 0.643 nm. E, View of all three longitudinal faces looking from the N terminus. F, View of the first and third faces from the N terminus. G, View of the first and second faces from the C terminus. Blue, Pro; red, Ser; light blue, Arg; gray, Leu. Backbone atoms are omitted for simplification. Images generated using DeepView, version 3.7 (http://www.expasy.org/spdbv).