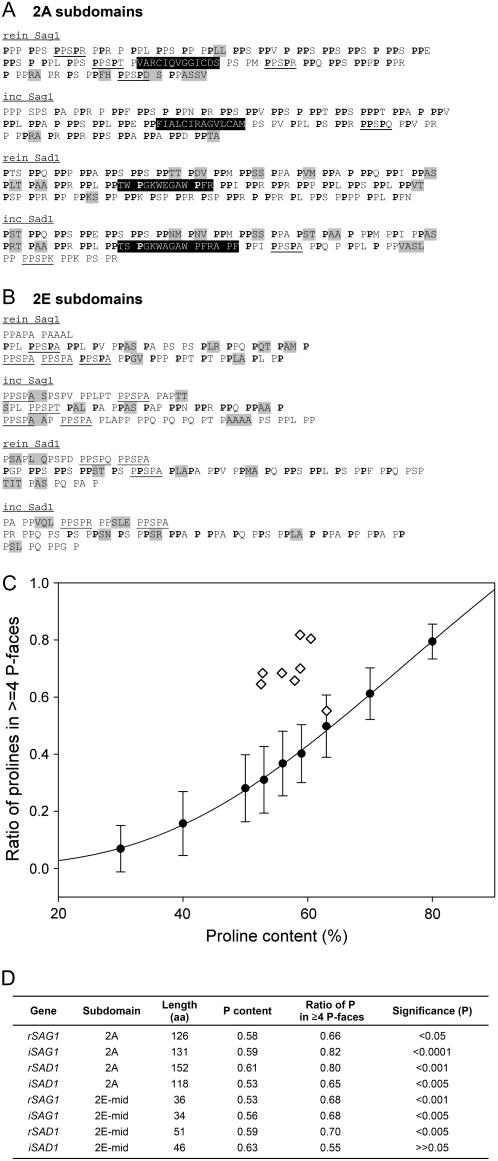
Figure 5.
Analysis of 2A and 2E shaft subdomains. A, 2A subdomains. PPX units are set off by spaces; light shading denotes guest sequences; dark shading denotes block interruptions; occasional PPSPX motifs are underlined. Boldfaced Pro residues participate in forming longitudinal face modules of ≥4 P. B, 2E subdomains, each displayed in three rows that correspond to within mating-type alignments (Supplemental Fig. S1). The first rows contain degenerate PPSPX residues; the third rows are also variable. The middle rows contain Pro residues (boldfaced) that participate forming longitudinal face modules of ≥4 P; these are considered in the simulation analysis. Other symbols as in Figure 5A. C and D, Simulation analysis. One hundred random protein domains, 100 amino acids in length, were generated with Pro content from 30% to 80%. Each sample was analyzed for its endowment of longitudinal faces containing ≥4 P; results plotted as circles in C. Diamonds in C show values for the actual sequences listed in D; P-value calculations (D) based on probability distribution of longitudinal Pro profiles in 100 randomly shuffled domains with the same Pro content.