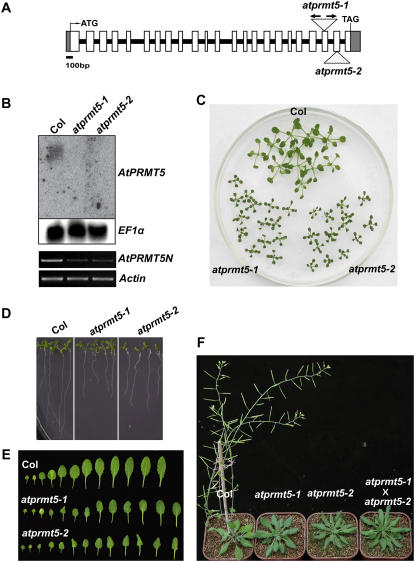
Figure 4.
AtPRMT5 gene structure, expression, and pleiotropic phenotypes in atprmt5-1 and atprmt5-2 mutants. A, AtPRMT5 gene structure and the T-DNA insertion sites of SALK lines. White boxes and gray boxes represent exons and 5′ or 3′ untranslated region, respectively. Lines mean introns. The T-DNA insertion sites in each mutant are indicated by triangles. In atprmt5-1 mutant, arrows represent two inverted T-DNA insertions. B, AtPRMT5 full-length but not the N-terminal transcript is absent in the atprmt5 mutants. Top sections: The total RNA from seedlings with four to five rosette leaves of atprmt5 mutants and wild-type Col is probed by RNA blot with full-length coding sequence of AtPRMT5. EF1α gene is used as a control for constitutive expression. Bottom sections: RT-PCR analyses were performed for the N-terminally expressed AtPRMT5 (AtPRMT5N) in wild-type Col and atprmt5-1 and atprmt5-2 mutants. Equal amounts of cDNAs were determined by RT-PCR at the Actin locus. C to F, Pleiotropic phenotypes of atprmt5 mutants. Growth retardation of young seedling leaves at 12 d (C) and primary roots at 9 d (D). E, Comparison of rosette leaves between wild-type Col and atprmt5 plants. F, atprmt5 mutants are late flowering. Plants shown here are 8 weeks old grown at 23°C under LD. aprmt5-1 × aprmt5-2 means F1 progeny from the crosses between aprmt5-1 and aprmt5-2.