Abstract
Prolate or c2-like phages are a large homologous group of viruses that infect the bacterium Lactococcus lactis. In a collection of 122 prolate phages, three distinct, non-cross-hybridizing groups of origins of DNA replication were found. The nonconserved sequence was confined to the template for an untranslated transcript, PE1-T, 300 to 400 nucleotides in length, while the flanking sequences were conserved. All three origin types, despite the low sequence homology, have the same functional characteristics: they express abundant PE1-T transcripts and can function as origins of plasmid replication in the absence of phage proteins. Using chimeric constructs, we showed that hybrids of two nonhomologous origin sequences failed to function as replication origins, suggesting that preservation of a particular secondary structure of the PE1-T transcript is required for replication. This is the first systematic survey of the sequence and function of origins of replication in a group of lactococcal phages.
Lactococcus lactis is widely used in the production of fermented products by the dairy and food industries. Large numbers of bacteriophages infect this bacterium, causing the lysis of the host and fermentation failure. The most frequently encountered are two species of small phages with isometric heads and B1 morphology, named 936 and P335, and one species of phages with prolate heads and a B2 morphology, named c2 (32). Each of the species comprises numerous homologous phages.
A number of host defense mechanisms against lactococcal phages have been discovered (reviewed in reference 12). Collectively, these mechanisms affect every step of the phage life cycle. Individual mechanisms may have a narrow range, affecting just a few strains of a single phage species, or broad range, affecting strains of several phage species. Under the pressure of the host defense mechanisms, phages may mutate or recombine with the chromosome or resident plasmids to produce resistant progeny (5, 15, 20, 36, 51). The interplay between the host defense mechanisms and phage evasion strategies is most likely a strong evolutionary driving force in the evolution of both.
Although the adsorption proteins are usually the most divergent part of the genome within a group of interbreeding (recombining) phage, variation of the DNA replication origin has also been described (7, 14, 30, 45, 51). In the lactococcal phage P335 species, it has been shown that under the selective pressure of the host defense mechanisms AbiC or AbiK, recombination can lead to the replacement of the origin of replication by a dissimilar prophage-derived sequence from the bacterial chromosome (5, 15). Recombination occurred via sequences of high identity flanking the divergent regions (5). This kind of module acquisition or shuffling is well noted in the bacteriophage world. A canonic example is the lambdoid phage family, in which the modules consisting of origin of replication, integration, lysogeny, and immunity control vary among the members. Despite variation in sequences, the functional organization of the whole module is preserved (7).
Prolate phages have the smallest genome among the lactococcal phages. The 20- to 22-kbp double-strand DNA genomes code for two blocks of divergently oriented open reading frames (ORFs), separated by a noncoding region. In phage c2, this intergenic fragment containing the early promoter 1 (PE1) and downstream 307 bp that serve as a template for an untranslated transcript (PE1-T) is sufficient to support plasmid replication in L. lactis in the absence of phage proteins (53). The PE1-T template sequence was confirmed as the origin of c2 replication by two-dimensional agarose gel electrophoresis of the replicative intermediates (6).
Two examples of transcript-mediated initiation of replication are phage T4 and plasmid ColE1. In the phage T4 middle origin, it was proposed that transcription from a middle-mode promoter followed by the formation of an RNA-DNA hybrid liberates the nontemplate DNA strand for primosome assembly and initiation of replication (9). In this case, phage proteins are only required for modification of RNA polymerase to enable transcription from the middle-mode phage promoter (9). In ColE1 and a few other plasmids in which replication is independent of plasmid-encoded initiator proteins, high homology of the sequences (80 to 90%) is confined to the template of a transcript at the origin of replication, called RNA II, required for initiation of replication (49). During transcription, the nascent ColE1 transcript forms an obligatory DNA-RNA hybrid that is processed by RNase H to form the mature RNA II. RNA II serves as a primer for polymerase I (PolI), which initiates replication (23). Particular secondary structure of the transcript is necessary for the ColE1 RNA-DNA persistent hybrid to form (33, 34).
The two prolate phage genomes (c2 and bIL67) that have been sequenced to date have an overall sequence identity of 80% (29, 47). Apart from several DNA insertions and/or deletions (“indels”), the highest diversity was detected in the putative adsorption protein gene (L10), minor coat protein gene (L16) and the 300 to 400 bp downstream of the PE1 promoter within the origin (29). The nonconserved region of the origin begins at position +16 of the PE1-T template and extends over most of its length, ending 25 nucleotides (nt) 5′ to the −35 box of the downstream early promoter, PE2. The bIL67 PE1-T template is longer by 72 bp than that of c2, and the nucleotide identity between the PE1-T template sequences is only 19%.
We report here that there are three unrelated types of PE1-T template sequences in a survey of 122 prolate phages. All three origin types are functionally analogous in that they serve as origins of replication in a plasmid model system in the absence of phage proteins and express abundant PE1-T transcripts. Two chimeric origins were nonfunctional in the plasmid system, suggesting that the secondary structure of PE1-T, rather than putative common sequence motifs, is essential for origin function. Based on searches of the GenBank database, we propose that diversity of the prolate phage origins was generated by horizontal transfer from other phage, plasmids or the lactococcal chromosome by recombination via microhomologous sequences.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Bacterial and phage strains used in this study are described in Table 1. L. lactis strains were grown in M17 medium supplemented with 0.5% (wt/vol) glucose at 30°C (50). For selection of L. lactis carrying plasmids pVA891, pTRKL2, and derivatives thereof, erythromycin (5 μg/ml) was added to the medium. Escherichia coli strain ER2206 was propagated on Luria-Bertani (LB) broth or brain heart infusion broth at 37°C. Antibiotics ampicillin and chloramphenicol were added to LB at 200 and 25 μg/ml, respectively, and erythromycin was added to brain heart infusion broth at 150 μg/ml as required.
TABLE 1.
Strains, plasmids, and bacteriophages used in this study
| Strain, phage, or plasmid | Descriptiona | Reference(s) and/or source |
|---|---|---|
| Strains | ||
| E. coli ER2206 | endA1 thi1 supE44 mcr67 (mrcA) (mrcBC-hsdRMS-mrr) 114::IS10 (lac)U169/F′ proAB laqIqZM15 Tn10 | New England Biolabs |
| L. lactis | ||
| MG1363 | Plasmid-free strain and prophage-cured derivative of NCDO 712 | 17 |
| Uvs62 | MG1363polA | 16 |
| IL1403 | Plasmid-free strain | 11 |
| c6 | 41 | |
| 112 | NZ dairy plant isolate | |
| AM1 | NZ dairy plant isolate | |
| 2282 | NZ dairy plant isolate | |
| Bacteriophages | ||
| c2 | MG1363 and IL1403 | 25, 35, 39 |
| c6A | c6 and IL1403 | 25, 41 |
| bIL67 | IL1403 and c6 (at 37°C only) | 47 |
| 923 | 112, AM1, MG1363 (at 37°C only), IL1403 (at 37°C only) | NZ dairy plant isolate (24) |
| 943 | AM1, 112, MG1363 (at 37°C only) | NZ dairy plant isolate, 1995 |
| 5440 | 2282 | NZ dairy plant isolate, 1995 |
| 5447 | 2282 | NZ dairy plant isolate, 1995 |
| 5469 | 2282, MG1363, IL1403 (at 37°C only), 112, AM1 | NZ dairy plant isolate, 1995 |
| Plasmids | ||
| pGEM-3Zf | Ampr; T7 and SP6 promoters | Promega |
| pGEMc2 | Ampr; c2 ori fragment cloned into pGEM-3Zf | This study |
| pGEMbIL | Ampr; bIL67 ori fragment cloned into pGEM-3Zf | This study |
| pGEM923 | Ampr; 923 ori fragment cloned into pGEM-3Zf | This study |
| pUCΩKm-2 | pUC carrying the Streptococcus faecalis Ω element | 38 |
| pTRKL2 | Eryr shuttle vector | 37 |
| pVAΩ | pVA891 carrying the transcriptional terminator of the Streptococcus faecalis Ω element | This study |
| pVA891 | Eryr Cmr; low-copy-number vector in E. coli; Eryr is expressed in gram-positive bacteria | 31 |
| pLP203 | Eryr; 369-bp ori fragment from c2 cloned into pVA891 | This study |
| pO923 | Eryr; 434-bp ori fragment from 923 cloned into pVA891 | This study |
| pObIL | Eryr; 444-bp ori fragment from bIL67 followed by the terminator from the Ω element cloned into pVA891 | This study |
| pSc2-923 | Eryr; 418-bp chimeric ori-fragment cloned into pVA891 | This study |
| pS923-c2 | Eryr; 415-bp chimeric ori-fragment cloned into pVA891 | This study |
| pMc2-923 | Eryr; 434-bp chimeric ori-fragment cloned into pVA891 | This study |
| pUC19-ori | Ampr; 369-bp ori fragment from c2 cloned into pUC19 vector | 53 |
For strains, description concerns phenotypes. For bacteriophages, description concerns host specificity. For plasmids, description concerns characteristics or further modifications.
Bacteriophage propagation.
Preparation of phage stocks was carried out in liquid medium or by plate lysis (24). When required, phages were concentrated by centrifugation at 40,000 × g for 2 h in a Sorvall centrifuge, or by precipitation with polyethylene glycol (24).
Recombinant DNA methods.
Basic cloning procedures in E. coli were as described previously (44). For purification of PCR products and for isolation of DNA from agarose gels, commercial DNA purification kits from Roche Molecular Biochemicals, Qiagen and Invitrogen were used according to the manufacturer's instructions. L. lactis electrocompetent cells were prepared using the glycine method (22). Typically, L. lactis cells were electroporated with 500 ng of plasmid DNA dissolved in water (40). Plasmid DNA was isolated from L. lactis by alkaline lysis (3). The plasmids were resolved by agarose gel electrophoresis (0.8%). To reveal the pObIL bands from the IL1403 transformant, the Southern blot was carried out using the pObIL plasmid as a probe and the ECL detection system (Amersham-Pharmacia) according to the manufacturer's instructions. The DNA from polyethylene glycol-precipitated phages was isolated using the Qiagen λ phage DNA purification kit (Midi) according to the manufacturer's instructions.
Construction of plasmids.
Plasmids that served as templates for synthesis of antisense probes were derivatives of pGEM-3Zf− (Promega). The inserts were amplified by PCR using appropriate primers and DNA from phage particles as templates. All primers (Table 2) contained recognition sites for appropriate restriction endonucleases: BamHI (forward primers) and EcoRI (reverse primers). The forward primers were: ts/+1 (c2), JR169 (bIL67), and JR170 (923). The reverse primer for all three clones was JR171. Amplified fragments were cloned into EcoRI and BamHI sites of the vector. The resulting template plasmids, named pGEMc2, pGEMbIL, and pGEM923, were used for synthesis of c2, bIL67, and 923 antisense probes, respectively, as described below.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′→3′)a | Specificity | Restriction site(s) |
|---|---|---|---|
| AS termin | TCGGAATTCGCTTGTAAACCGTTTTGTGAA | Ω element | EcoRI |
| JR120 | CTTTGTTAATTGCTTTGATGTCGTC | Conserved | |
| JR124 | CGATAAAATAACCGTTACAATTAGCC | Conserved | |
| JR139 | CCTTGAGTTGTCTATGGTTGCTAA | bIL67 | |
| JR166 | GCTGACATTAWCCAATAAA | Conserved | |
| JR167 | GATTATGGTATTATTATAGCA | Conserved | |
| JR169 | CGGGATCCATCAAAAATTTAACGACTGTTA | bIL67 | BamHI |
| JR170 | CGGGATCCAACGTATGTTATAATATAAATA | 923 | BamHI |
| JR171 | CGAATTCGCTGACATTATCCAATAAA | Conserved | EcoRI |
| JR172 | ACGCAAACGCAGTTTTTATCC | c2 | |
| JR173 | CTAAGGCTTGTCTGATGTCTT | c2 | |
| JR174 | GCCCTTGCCTTTTTGGTTAAG | bIL67 | |
| JR177 | GCGAGGCGAAAGCCTATGAA | 923 | |
| JR178 | TCGGAATTCCCATGGCTTATGTTTTTGACCCTAA | bIL67 | NcoI |
| JR179 | TCGGAATTCTATGTTACTCTTTAATTACAA | 923 | EcoRI |
| JR180 | TCGGAATTCCCATGGCTTGTGTTTTTTACCCT | 923, c2 | NcoI |
| JR184 | TATTATAACATACGTTTT | Conserved | |
| JR185 | AAAACGTATGTTATAATA | Conserved | |
| JR203 | GTAGAGATTCTGATAAGGTAG | 923 | |
| JR204 | CATGCCATGGAACTGCAGTGGATGACCTTTTGAATGAACC | Ω element | NcoI/PstI |
| JR205 | AAAACTGCAGTAGTTACTTTATTTTAGACAA | bIL67 | PstI |
| JR206 | CAACCTGTCCACTTcttatttaactttgcc | 923/c2 (chimeric) | |
| JR207 | gttaaataagAAGTTGGACAGGTTGTGAG | c2/923 (chimeric) | |
| JR221 | GTTGTGAGTaagttaaataagaagttac | 923/c2 (chimeric) | |
| JR222 | cttatttaacttACTCACAACCTGTCCAC | c2/923 (chimeric) | |
| JR223 | CATGCCATGGGTCGACCTTGTGTTTTTTACCCT | 923 | NcoI |
| Latedel | AAAGAATTCCTTGTATTTTTGACCCTG | c2 | EcoRI |
| pUC19 −20b | GTAAAACGACGGCCAGT | M13/pUC | |
| ts/+1 | CGGGATCCATAAAAATTGAATACGCC | c2 | BamHI |
In chimeric oligonucleotides, uppercase and lowercase letters represent, respectively, 923 and c2 sequences. Restriction sites are shown in italics.
From New England Biolabs.
To assay the origin function of c2, bIL67, and 923 PE1 transcripts, ori sequences containing the PE1 promoter starting from position −62 and downstream template sequence extending nearly to the −35 box of the subsequent downstream early promoter, PE2, were cloned into the origin probe vector pVA891 (31). The ends of the template fragments were 20 nt (c2 and bIL67) and 9 nt (923) upstream of the −35 box of PE2 and therefore did not carry any promoter sequences of PE2. The c2 ori fragment was amplified using primers pUC19 −20 (New England Biolabs) and Latedel and plasmid pUC-ori (53) as template. The resulting fragment was cleaved with EcoRI and cloned into the EcoRI site of pVA891, and the plasmid was named pLP203. The 923 ori fragment was amplified using the primers JR180, JR179, and 923 phage as a template. The amplified PCR product was cloned into the NcoI and EcoRI sites of pVA891, and the plasmid was named pO923.
The bIL67 ori fragment was cloned into the modified pVA891, named pVAΩ, which carried the transcriptional and translational terminator from the Streptococcus faecalis Ω element cloned between the NcoI and EcoRI sites. A unique PstI site was engineered, so that the inserts could be directionally cloned into PstI/NcoI-cut vector upstream of the terminator, which ends the transcription and prevents read-through into the vector. The terminator was on a 130 bp insert created by PCR, using primers JR204, AS termin, and plasmid pUC4Ωkm-2 (38) as template. The bIL67 ori fragment was amplified using primers JR178, JR205, and bIL67 phage as template, and was inserted into the NcoI and PstI sites of vector pVAΩ. The resulting plasmid was named pObIL.
Chimeric ori inserts for cloning into pVA891 were composed of portions of the ori regions of c2 and 923 phage. They were constructed by overlap extension PCR, carried out in two steps. In the first step, origin halves, each from different phage, were amplified separately. The reverse primer for amplification of the 5′ half and the forward primer for amplification of the 3′ half were chimeric and complementary to the other. Thus, the PCR products amplified using the chimeric primers carried complementary ends. In the second step, an overlap extension PCR was used to join the two halves into a chimeric ori. The equimolar amounts of products of the first round of PCR served as templates. The primers corresponded to 5′ and 3′ ends of the chimera and included the restriction sites, EcoRI and NcoI, respectively, to allow cloning into the EcoRI and NcoI sites of the vector pVA891. Three chimeric ori constructs were made. The first construct, pSc2-923, carried the c2/923 chimeric ori, composed of the PE1 promoter and 5′ half of c2 ori (62 + 172 bp) and 3′ half of the 923 ori (184 bp). Primers used for amplification of the c2 fragment were JR180 and JR206, using c2 phage as template. For amplification of the 923 phage fragment, primers JR207, JR179, and 923 phage were used as templates. The second round of PCR was carried out using primers JR180 and JR179 and the two products of the first round of PCR as templates. The second construct, pS923-c2, carried a 923/c2 chimeric ori consisting of the PE1 promoter and 5′ half of 923 ori (62 + 200 bp) and the 3′ half of the c2 ori (153 bp). Primers used for amplification of the 923 fragment were JR223 and JR222 and the template was 923 phage. For amplification of the c2 fragment, primers were JR221 and pUC19 −20 and the template was pUC-ori plasmid (53). The chimeric insert was amplified in the second round of PCR, using primers JR223 and pUC19 −20, and the two products of the first round of PCR as templates. In the third chimeric construct, pMc2-923, the junction of the chimera was at the PE1 promoter. Thus, although the ori was chimeric, the entire template sequence of the PE1-T template sequence was derived from a single phage. The PE1 promoter from position −62 to −27 was from c2, followed by identity of 20 bp, and a portion of 923 from −7 to + 367. The c2 portion of the insert was amplified using primers JR180, JR184 and c2 phage as template. The 923 portion of insert was amplified using primers JR185, JR179, and 923 phage as templates. The chimeric insert was amplified in the second round of PCR, using primers JR180, JR179, and the two products of the first round of PCR as templates.
Sequencing of the phage origins of replication.
Origin fragments equivalent to the genomic c2 sequence from coordinates 6353 to 7685 (29) were sequenced in 6 prolate phage: c6A, 923, 943, 5440, 5447, and 5469. The ori fragments were first amplified by PCR, using primers JR120, JR124 and DNA from phage particles as templates. To minimize the error rate, proof-reading polymerase Pwo (Roche Molecular Biochemicals) was used. As an additional precaution, each preparative PCR was divided in four PCR tubes prior to cycling. After the PCR, the four reactions were combined for use in the subsequent purification and sequencing. The sequencing was carried out using the BigDye mix (Applied Biosystems) at Alan Wilson Centre Genome Service (Massey University). Both strands were sequenced by primer walking, each with at least twofold redundancy. Sequences were assembled and analyzed using the GeneWorks program (Oxford Molecular-Accelrys) and BioEdit (Tom Hall, North Carolina State University, Raleigh). The GenBank accession numbers for the phage origin sequences are AF522295 for 923, AY129505 for 943, AY129506 for 5440, AY129507 for 5447, AY129508 for 5469 and AY129509 for c6A.
Survey of the ori regions from industrial prolate phage isolates.
ori regions from 116 prolate phage isolates from the Fonterra Research Centre (formerly New Zealand Dairy Research Institute) collection were amplified using the primers complementary to the conserved sequences flanking the divergent PE1-transcript template sequence, JR167, JR166 and DNA from phage particles as templates. The PCR products were arrayed in triplicate on a Nitrocellulose filter using Multi-Blot replicator (V&P Scientific, Inc.) (see Fig. 3), or separated by agarose gel electrophoresis and transferred to the Nitrocellulose membrane (not shown). The types of origin were distinguished using type-specific probes derived from the nonconserved PE1-T template region by PCR. The c2 probe was amplified using primers JR173 and JR172 with c2 phage as a template; the bIL67 probe was amplified using primers JR174 and JR139 with bIL67 phage as a template; and 923 probe was amplified using primers JR177 and JR203 with 923 phage as a template. The hybridization was detected using the ECL system (Amersham-Pharmacia).
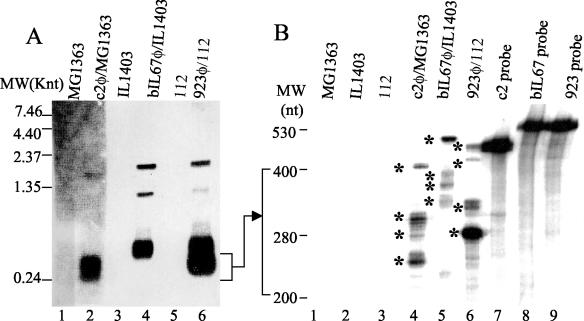
FIG. 3.
PE1 transcripts of c2, bIL67 and 923 phage. (A) Northern blot of RNA resolved by denaturing agarose gel electrophoresis. Each lane was loaded with 20 μg RNA. Lanes 1 (MG1363) and 2 (c2 phage-infected MG1363) were probed with a c2 probe; lanes 3 (IL1403) and 4 (bIL67 phage-infected IL1403) with a bIL67 probe; lanes 5 (112) and 6 (923 phage-infected 112), with a 923 probe. The probes were ori-specific PCR fragments derived from nonconserved PE1 template sequences. (B) RNase protection of PE1 transcripts, resolved by high resolution acrylamide gel-electrophoresis. Lanes 1 to 6, RNase protection reactions of the RNA from MG1363 (lane 1), IL1403 (lane 2), 112 (lane 3), c2 phage-infected MG1363 (lane 4), bIL67 phage-infected IL1403 (lane 5), and 923 phage-infected 112 (lane 6); lanes 1 and 4, reactions with the c2 probe; lanes 2 and 5, the bIL67 probe; lanes 3 and 6, 923 probe. Lanes 1, 2, 4, and 5 contain 26 μg of RNA per reaction. Lanes 3 and 6 contain 13 μg of RNA per reaction. Lanes 7, 8, and 9 contain probes only (lane 7, c2; lane 8, bIL67; lane 9, 923). Asterisks indicate major PE1 transcripts. c2, bIL67, and 923 ori-specific probes were synthesized by in vitro transcription using plasmids pGEMc2, pGEMbIL, and pGEM923 (described in Materials and Methods) as templates.
RNA purification and analysis.
For the phage RNA analyses, cells were infected at an optical density of 0.1 and a multiplicity of infection of 5 and harvested 15 min after infection, the time point at which the PE1 transcript accumulates to a high level (28). RNA isolation was carried out as described by Lubbers et al. (28), with the following modifications: instead of the hot phenol treatment, the cells were broken in cold phenol (4°C) using acid-washed glass beads (0.6 mm), at 4°C with continual vortexing for 10 min (2). The mixture was centrifuged at 4°C and the clear water phase was collected, extracted with chloroform, precipitated and resuspended in TE (10 mM Tris, 1 mM EDTA), pH 7.5. The samples were then treated with DNase I (Roche) to eliminate the remaining DNA, and subjected to another round of phenol-chloroform and precipitation as described (28). The concentration of RNA samples was determined spectrophotometrically and the quality of preparation examined by agarose electrophoresis (44).
RNase protection was carried out as previously described (44). Radioactively labeled antisense probes were generated in vitro using the Promega T7 riboprobe kit and [α-32P]CTP (Amersham-Pharmacia) according to the manufacturer's instructions. Templates for synthesis of antisense probes used to detect c2, bIL67, and 923 PE1 transcripts were pGEMc2, pGEMbIL67, and pGEM923, respectively. The antisense probes were complementary to the PE1-T template starting from + 1 position (c2 and bIL67) or −20 position (923), comprising the variable portion and extending 93 nt into the downstream conserved region. Thus, the probes detected the full length of the nonconserved region of PE1 transcripts and 93 nt of downstream conserved sequence. In addition to the phage sequences, all three probes carried at the 3′ end residual vector sequence, retained after restriction enzyme cleavage to linearize the template prior to the in vitro transcription.
RNase-protected fragments of radioactively labeled probes were separated by denaturing polyacrylamide gel electrophoresis on a large gel (40 cm). Size standards were used to allow accurate determination of the length of the RNA fragments: end-labeled low-molecular-weight RNA standard (Invitrogen) and a sequencing reaction generated using an end-labeled primer.
For Northern blots, RNA was separated using agarose-formamide gels (44), and detected using PCR-generated DNA probes and the ECL system (Amersham-Pharmacia) according to the manufacturer's instructions. The probes used for the Northern blots were the same as ones used for the Southern blot survey of the origins of replication of prolate phage isolates.
Modeling of the RNA secondary structures was performed using the program ALIFOLD (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi [21]).
RESULTS
Sequence diversity of the origin of replication in prolate phage.
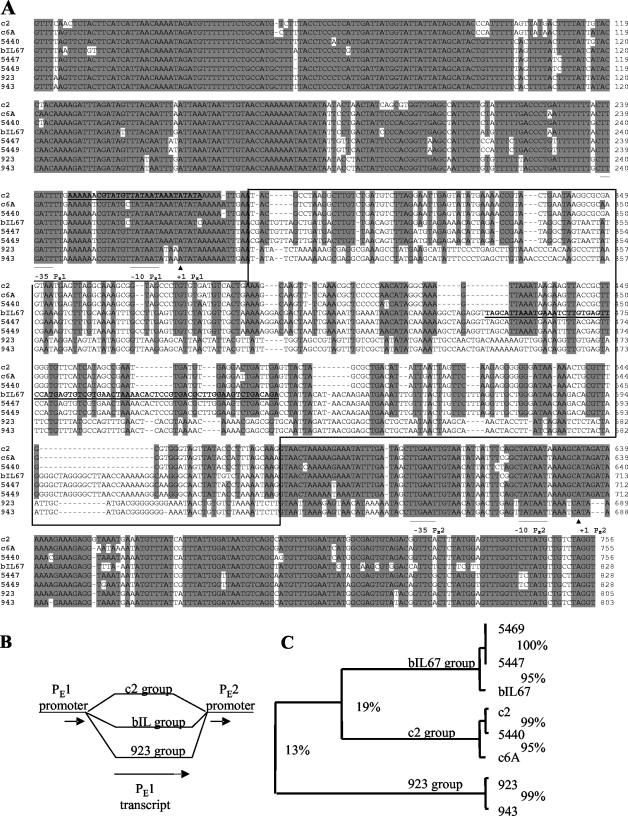
To determine the extent of variation of the PE1-transcript template segment and flanking sequences in prolate phages, the sequence of ca. 1,300 bp of that region was determined in six phage isolates (Fig. 1A). Five phages were isolates from New Zealand dairy plants: 923, 943 (both in 1975), 5440, 5447, and 5469 (all three in 1995). The sixth phage, c6A, was a 1960s isolate from Australia and has been an object of several publications (41-43). Alignment of the newly and previously sequenced phage origins revealed three groups of PE1-T templates, sharing only 13% identity. In contrast, the flanking sequences were conserved in all phages (Fig. 1A and B). The PE1-T template sequences of c2, c6A, and 5440 belonged to one group (c2 type ori); bIL67, 5447, and 5469 belonged to the second (bIL67 type ori); and 923 and 943 belonged to the third (923 type ori; Fig. 1C). Despite the sharp divergence in the PE1-T templates among the three types, within a group they were highly conserved, even more so than the flanking sequences. For example, overall identity of the 1,300-bp fragment within the c2 ori group phages (c2, c6A, and 5440) was 89%, while PE1-T template identity was 95%.
FIG. 1.
(A) Alignment of PE1 transcript template and flanking sequences. Sequences were aligned using the ClustalW multiple alignments program (52) within the BioEdit sequence analysis package (Tom Hall, North Carolina State University, Raleigh). The aligned sequences start at position 7294 and end at position 6540 of the c2 genomic sequence. The −35 and −10 boxes of PE1 and PE2 promoters are underlined, and the + 1 residues labeled with the symbol ▴ (28). The nonconserved portion of the alignment is boxed. The two highlighted sequences (in boldface type and underlined) showed significant identities with sequences from the NCBI nucleotide database: nt 247 to 275, 96% identity to the L. lactis IL1403 chromosome; nt 449 to 525, 88% identity to the small isometric phage bIL170; and nt 470 to 525 (contained within the preceding sequence), 92% identity to the plasmids pAW601 and pIL103. (B) Schematic representation of origin divergence. Single lines represent the conserved sequences and branched lines the divergent region; short arrows, promoters; long arrow, PE1 transcript template. Note that the c2 group PE1-T template is shorter than that of the bIL67 and 923 groups. The dotted portion of the arrow symbolizes the length difference. (C) Rectangular cladogram based on the ClustalW alignment of PE1 templates only (nonconserved portion of sequences, boxed in A). Numbers represent % identities between the phage and phage groups.
To examine whether prolate phages carry yet other types of ori, we surveyed origins of replication of the entire collection of prolate phage isolates of Fonterra Research Centre, which consists of 116 isolates collected systematically over the last 55 years from all major dairy plants in New Zealand. Identity of all prolate phages is verified by electron microscopy (L. Ward and A. Jarvis, unpublished data). Origin fragments containing the nonconserved PE1-T sequences were amplified by PCR, using primers complementary to the flanking conserved regions, and DNA from phage particles as template. The PCR-amplified fragments were analyzed by hybridization with three ori type-specific probes derived from the nonconserved PE1-T templates of representative phage, c2, bIL67, and 923 (Fig. 2). All PCR-amplified origins from prolate phage hybridized with one and only one of the three PE1-T probes, showing that there were only three types of ori present in the New Zealand prolate phages surveyed, and that there were no hybrid origins (composed of parts of two origin types). The c2 type ori was the most frequent (72 isolates), followed by bIL67 type (25 isolates) and 923 (19 isolates).
FIG. 2.
Survey of prolate phage origin types. The PCRs of the origin fragments of 116 prolate phage isolates from Fonterra Research Centre collection and 8 phage from which the ori sequence has been determined were arrayed in triplicate and each array was probed with a type-specific probe. (A) c2 probe; (B) bIL67 probe; (C) 923 probe. The PCRs of the sequenced origins are boxed. Type-specific probes were PCR fragments derived from the nonconserved PE1-T template sequences.
All ori types express PE1-T transcripts.
It has previously been shown that the c2 PE1 promoter expresses several short and abundant noncoding PE1-T transcripts, 250 to 360 nt in length (28). We compared the abundance and length of PE1-T transcripts in representative phage of all three ori types by Northern hybridization and RNase protection analyses of RNA isolated from phage-infected cells (Fig. 3). Northern hybridization revealed abundant short PE1 RNAs (260 to 500 nt) in all (c2, bIL67, and 923) phage-infected cells (Fig. 3A). In addition, one (c2) and two (bIL67 and 923) longer and less abundant RNA species were detected. Of those, 1.35- and 1.9-knt transcripts were common to bIL67 and 923, while the corresponding c2 transcript was 1.8 knt.
To determine the composition and length of the short PE1 transcript bands, RNase protection was carried out with two sets of probes: one longer probe set (described in the Materials and Methods section), extending beyond the nonconserved region (Fig. 3B), and a set of shorter probes (not shown). The products were resolved by polyacrylamide gel electrophoresis on high-resolution sequencing gels (Fig. 3B). The most striking feature was the presence of multiple bands, consistent with the appearance of a smear on Northern blots after the agarose gel electrophoresis (Fig. 3A) (27). The major protected bands were of different lengths in each phage, consistent with the diversity of the PE1 template sequences (Fig. 3B). The longest bands, nearly as long as the full-length probes (excluding the vector-derived and upstream sequences), were protected by long transcripts that extended beyond the end of the antisense probe. This is consistent with the presence of longer transcripts detected by Northern blotting (Fig. 3A).
The RNase protection data indicated that the estimated lengths of the following major PE1 RNAs from this experiment were as follows: c2, 260, 265, 280, 295, and >419 nt; bIL67, 330, 360, 400, and >489 nt; 923, 280, 325, 330, 440, and >466 nt. The dominant bIL67 transcript was longer than the probe, indicating that its 3′ end corresponds to the downstream conserved sequence.
All three types of PE1 RNA templates are replication origins in the absence of phage proteins.
It has recently been shown that the intergenic region of c2 phage (611 bp), carrying the promoter and nonconserved PE1-T template sequence, is sufficient to drive replication of the origin probe plasmid pVA891 in the absence of phage proteins (53). To examine whether the analogous regions of bIL67 and 923 could function as origins of plasmid replication, appropriate DNA fragments from the two phage were inserted into the origin-screening plasmid pVA891 (31), which lacks an origin function in L. lactis. Each insert started at the −62 position of PE1 and ended at the point of reestablishment of homology: +307 in c2, +382 in bIL67 and + 372 in 923 (Fig. 1). In the 923 type of ori, identity resumes 1 nt 5′ to the −35 box of the downstream promoter, PE2. In bIL67, the homology to c2 phage starts 26 nt 5′ to the −35 box of PE2 promoter (Fig. 1A).
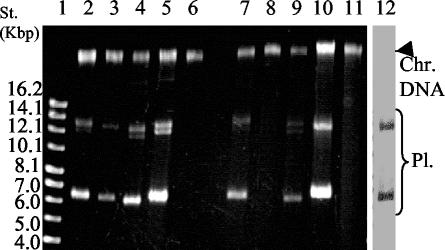
The three pVA891 derivatives, pLP203, pObIL, and pO923, were transformed into L. lactis, and their ability to replicate was examined. Because the major transcript of bIL67 PE1 extends beyond the end of the insert, the bIL67 construct carried a transcriptional terminator between the 3′ end of the insert and the vector to prevent transcriptional read-through into the vector sequence. As each of the three phages plate on a distinct cognate host strain, and it was possible that diversity of origins was important for host-specificity, each of the three hosts (MG1363, IL1403, and 112) was electroporated with all three plasmids. The transformation efficiencies were compared with a control plasmid, pTRKL2, which carries pAMβ1 origin of replication that is functional in a broad range of Gram-positive bacteria (37). All plasmids conferred Ery resistance to all strains, suggesting that they replicated in all strains. The IL1403 transformants that carried pLP203 and pO923 grew much faster than those carrying pObIL (24 h to form a colony on solid media compared to 72 h for pObIL transformants). When transformants were analyzed for plasmid content (Fig. 4), the relatively small amount of plasmid recovered per cell from pObIL transformants compared to the other ori plasmids. The pObil plasmid bands isolated from the IL1403 transformant were detectable only by Southern blotting (Fig. 4, lane 12). Probe, the full-length pObIL plasmid, did not hybridize to the chromosomal band, indicating that the plasmid was not integrated into the chromosome. Therefore, the low yield was most likely due to relatively inefficient replication of this plasmid rather then from integration into the chromosome. The low efficiency of pObil replication in IL1403 might be due to the truncation of the bIL67 PE1-T in pObIL, since the major bIL67 transcript in the phage extends into the downstream conserved region (Fig. 3B). However, the longer fragment proved recalcitrant to cloning and hence could not be used in this experiment.
FIG. 4.
ori plasmid preparation from L. lactis strains MG1363 and IL1403. ori constructs are derivatives of the origin-less plasmid pVA891 (31), each harboring a prolate phage promoter PE1 (61 bp) and downstream PE1-T template sequence. Plasmid pLP203 carries the c2 phage ori (369 bp), pObIL carries the bIL67 ori (444 bp); pO923 carries the 923 ori, (434 bp). pTRKL2 is a lactococcal plasmid with pAMβ1 origin of replication (37). Lanes: 1, supercoiled DNA standard; 2 to 11, EtBr-stained agarose gel electrophoresis of the plasmid preparations: 2, pLP203; 3, pObIL; 4, pO923; 5, pTRKL2, all isolated from MG1363; 7, pLP203; 8, pObIL; 9, pO923; 10, pTRKL2, all isolated from IL1403. Lanes 6 and 11 are the preparations of the host strains MG1363 and IL1403 included to indicate residual chromosomal DNA in the samples. Lane 12, Southern blot of the pObIL plasmid preparation from IL1403 transformant. The blot was probed with the full-length pObIL plasmid. Each lane was loaded with 1/20 volume of a plasmid prep from 7 optical density units (at 600 nm) of late exponential culture. Abbreviations: Pl., plasmid DNA bands; Chr. DNA, chromosomal DNA band.
No ribosome-binding sites or ORFs longer than 36 bp were encoded by PE1-T template of c2 (28), bIL67 (47), and the six prolate phages sequenced in this study. Thus, it may be inferred that the prolate phage origins are functional in the absence of phage proteins. Accordingly, the prolate origins of replication do not exhibit the phage resistance phenotype (per), even when carried on high copy number plasmids (J. Rakonjac and A. Schiemann, unpublished data).
Chimeric PE1-T template sequences do not function as origins of replication.
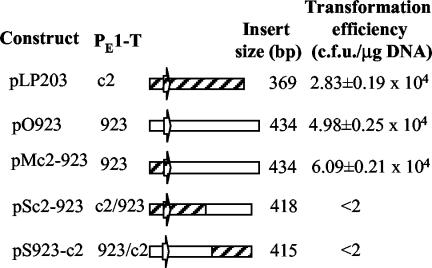
The small number (three) of PE1-T variants found in prolate phage population, and the high conservation of PE1-T templates within a single ori type, suggested that the sequence integrity of a PE1-T transcript might be critical for origin function. Origin function might be dependent on PE1-transcript secondary structure, primary sequence motifs, or both. To distinguish between these possibilities, we constructed two chimeric origins: one was composed of the promoter and 5′ half of the c2 PE1-T template fused to the 3′ half of 923 PE1-T template (pSc2-923), and the other was the reverse, promoter and 5′ half from 923 and 3′ half from c2 (pS923-c2). As a control, a chimera that contained the complete PE1 coding sequence from c2, but the promoter from 923 phage (pMc2-923) was constructed (Fig. 5).
FIG. 5.
Schematic representation of chimeric ori constructs and their ability to support plasmid replication. Diagonally hatched, c2 sequences; white, 923 sequences. Block arrow, PE1 promoter. Transformation efficiency was tested using strain MG1363 as a recipient for electroporation.
Neither construct with the chimeric PE1-T template could drive replication of pVA891. In contrast, the control chimera was functional as a plasmid origin. This suggests that the integrity and perhaps conserved secondary structure of the PE1 transcript is important for replication.
Secondary structures are conserved within each ori type.
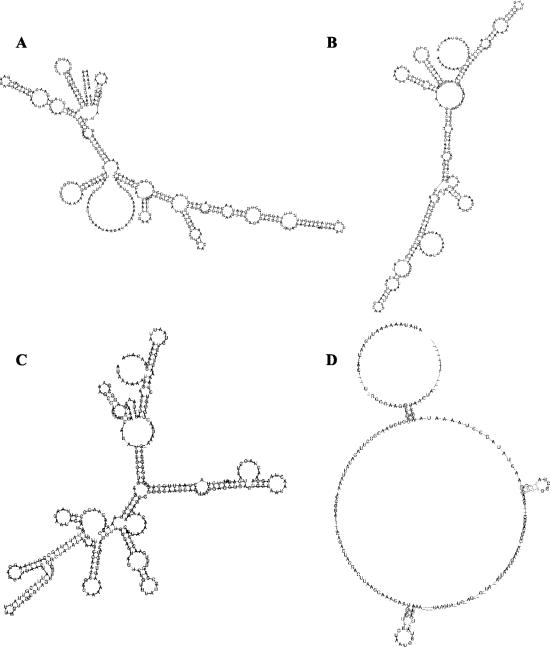
To assess the relationship between the secondary structures of the PE1 transcripts, ClustalW alignments of each of the three types of ori PE1 transcripts were subjected to the modeling of the secondary structures using the program ALIFOLD (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi) (21). The predicted structures suggest that most of the nucleotide differences among the origins of the same type were conserved (Fig. 6A, B, and C). Also apparent was that the predicted secondary structures of the three types of PE1 transcripts were different. When the alignment all sequenced origins was subjected to the ALIFOLD analysis, very few nucleotides showed conserved changes, confirming the absence of conservation of the secondary structure among the three origin types (Fig. 6D). The lack of conservation of secondary structure is consistent with the lack of functionality of the chimeric origins.
FIG. 6.
Secondary structure predictions of the aligned PE1 transcripts using the program ALIFOLD (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi) (21). (A) c2 type; (B) bIL67 type, (C) 923 type, (D) all sequenced origins.
Host requirements for prolate phage replication.
PolI is required for initiation of replication of ColE1 plasmid, where the origin transcript RNA II serves as a primer for this replicase. To determine whether the PolI is required for prolate phage replication, phage with c2 and 923 origins were plated on the polA mutant of MG1363 (16). These phages plated on polA mutant with the 100% efficiency relative to the parent MG1363 showing that PolI is not required for the initiation of replication.
BLAST searches for sequences with high identities to PE1-T templates and flanking sequences.
Variants of the prolate phage PE1-T templates could have been acquired during evolution by horizontal transfer, through recombination with other phage, plasmids or chromosome, via the short sequences of high identity. To look for sequence identities with other organisms, we carried out BLAST searches (1) using the PE1-T template sequences of c2, bIL67, and 923 phage as queries. No sequences significantly similar to the c2 PE1-T template were found in the GenBank nonredundant database, other than that of a prolate phage of the same origin type (46), nor were there any significant sequence identities to the 923 PE1-T sequence. Interestingly, the bIL67 PE1-T template shared a region of homology, 76 nt in length and 88% identity, with the intergenic sequence of a small isometric phage of the 936 species, bIL170 (positions 28949 to 29025) (13). Within this bIL67 sequence, a 57-nt stretch also shared homology to a sequence 480 nt upstream of the −35 box of the repB gene near the replication origin of plasmids pIL103 and pAW601 (92% identity) (18, 48; N. N. Matvienko, A. Madsen, and J. Josephsen, unpublished data [NCBI accession no. AJ132009]). Another BLAST search was carried out, with the sequence that included conserved sequences (60 nt on either side) flanking the divergent PE1 template. This search detected identity of the PE1 promoter of the c2 phage to L. lactis strain IL1403 genomic sequence (4), 28 nt long with 96% identity (nt 7399 to 7426; section 185). The identity is overlapping with the promoter of ORF coding for the putative phenolic acid decarboxylase (position −44 to −16). The prolate phage origin sequences with the above homologies were highlighted (bold and underlined) in Fig. 1A.
DISCUSSION
The prolate phage c2 origin of replication is functional in the absence of phage proteins (53). There have been no other reports of a lactococcal phage origin that drives plasmid replication in the complete absence of any phage protein, although the protein requirement is unclear for the phage sk1 (936 species) (10). The sk1 origin plasmid construct tested included an incomplete ORF, and the involvement of this ORF in replication has not been determined (10). We have now analyzed the requirements for replication of prolate phages other than c2 and showed that prolate phages have at least three types of PE1 RNA template sequences, which we have named the c2, 923, and bIL67 types, after the phages representing each group. These sequences shared only 13% identity among eight prolate phages. An exhaustive survey of the entire prolate phage isolate collection from New Zealand dairy plants with different geographical and temporal histories showed that there were only three types of PE1 RNA template sequences in New Zealand isolates over the last 50 years.
The region of sequence divergence of the prolate phage origin extended from the +16 nucleotide of the PE1-T template sequence to the −35 box of the next downstream early promoter, PE2. The flanking regions, including the PE1 and PE2 promoters, were highly conserved. BLAST searches using the prolate phage PE1-T template variable and the flanking conserved sequences as queries detected short highly identical sequences in phage, plasmid and chromosome. Thus, it is possible that new PE1-T templates in prolate phages could have been acquired by recombination from other lytic phages, resident plasmids or the lactococcal chromosome via the short highly homologous sequences (microhomologies), as proposed by Cambpell and Botstein for lambdoid phages (8). Similarly to previous reports of recombination in lactococcal phages of the P335 species (5, 15, 36), acquisition of a new origin in the prolate phages could have been selected for in hosts that carried a putative defense mechanism(s) targeting the origin of a particular type. An extensive cross-plating matrix analysis would be required to detect such strains in our phage/host strain collection. A small-scale experiment, with six host strains and corresponding phages of which the ori sequences were determined in this study did not detect any such strain (data not shown).
Despite the lack of conservation of their sequences, all three types of minimal origins served as templates for transcription of untranslated PE1 RNAs. The Northern blot and RNase protection experiments detected multiple PE1 RNAs in cells infected with phages representing the three ori types, c2, bIL67, and 923. Each type produced a unique pattern of PE1 RNAs, in accordance with different lengths and divergent sequences of the PE1-T templates. Sequence analysis did not detect appropriately positioned transcriptional terminators downstream of the 3′ ends of the PE1 RNAs. Therefore, it is possible that the mature PE1 RNAs are derived from longer precursor transcripts by host RNA-processing enzymes, such as RNase H, 3′-5′ exoribonucleases, RNase P, or RNase III (19, 26, 27).
Despite the lack of homology among the three types of PE1 RNAs, it was possible that putative short sequence motifs that they shared might be required for replication. This was tested by constructing precise c2-923 and 923-c2 chimeric origins. The chimeric constructs contained complete origin sequences—thus, any potential functionally important shared sequences would have been present—but they failed to replicate. Since specific sequence motifs are not sufficient for replication, the proper secondary structure is most likely required for function of PE1 transcripts. The transcript does not serve as a primer for PolI-dependent initiation of replication; thus, it is possible that, as in T4 phage, it serves to capture the coding strand and liberate the noncoding strand to allow assembly of the primosome and initiation of replication.
This study characterized RNAs expressed from replication origins in a family of homologous phages and suggested their role in replication. Further research will be required to determine the mechanism of processing, stabilization and initiation of replication by prolate phage PE1 RNAs.
Acknowledgments
We thank Marjorie Russel and Peter Model for critical reading of and suggestions about the manuscript, P. Duwat for the generous gift of the strain Uvs62, J. Gordon for plasmid pLP203, and Q. Deng for technical help.
This work was supported by the Marsden Fund of the Royal Society of New Zealand (grant MAU803). A. Schiemann was supported by a Massey University scholarship.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anba, J., E. Bidnenko, A. Hillier, D. Ehrlich, and M. C. Chopin. 1995. Characterization of the lactococcal abiD1 gene coding for phage abortive infection. J. Bacteriol. 177:3818-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 6.Callanan, M. J., P. W. O'Toole, M. W. Lubbers, and K. M. Polzin. 2001. Examination of lactococcal bacteriophage c2 DNA replication using two-dimensional agarose gel electrophoresis. Gene 278:101-106. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, A., and D. Botstein. 1983. Evolution of the lambdoid phages, p. 365-380. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 9.Carles-Kinch, K., and K. N. Kreuzer. 1997. RNA-DNA hybrid formation at a bacteriophage T4 replication origin. J. Mol. Biol. 266:915-926. [DOI] [PubMed] [Google Scholar]
- 10.Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 11.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 12.Coffey, A., and R. P. Ross. 2002. Bacteriophage-resistance systems in dairy starter strains: molecular analysis to application. Antonie Leeuwenhoek 82:303-321. [PubMed] [Google Scholar]
- 13.Crutz-Le Coq, A. M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 14.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 15.Durmaz, E., and T. R. Klaenhammer. 2000. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl. Environ. Microbiol. 66:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duwat, P., A. Cochu, S. D. Ehrlich, and A. Gruss. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier, M., and M.-C. Chopin. 1987. Plasmid-determined systems for restriction and modification activity and abortive infection in Streptococcus cremoris. Appl. Environ. Microbiol. 53:923-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gegenheimer, P., and D. Apirion. 1981. Processing of procaryotic ribonucleic acid. Microbiol. Rev. 451:502-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, C., L. A. Miller, and T. R. Klaenhammer. 1991. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 173:4363-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofacker, I. L., M. Fekete, and P. F. Stadler. 2002. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 319:1059-1066. [DOI] [PubMed] [Google Scholar]
- 22.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh, T., and J.-I. Tomizawa. 1980. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. 77:2450-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvis, A. W. 1984. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl. Environ. Microbiol. 47:343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keogh, B. P. 1973. Adsorption, latent period and burst size of phages of some strains of lactic streptococci. J. Dairy Res. 40:303-309. [Google Scholar]
- 26.Li, Z., and M. P. Deutscher. 1996. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell 86:503-512. [DOI] [PubMed] [Google Scholar]
- 27.Li, Z., S. Reimers, S. Pandit, and M. P. Deutscher. 2002. RNA quality control: degradation of defective transfer RNA. EMBO J. 21:1132-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lubbers, M. W., K. Schofield, N. R. Waterfield, and K. M. Polzin. 1998. Transcription analysis of the prolate-headed lactococcal bacteriophage c2. J. Bacteriol. 180:4487-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubbers, M. W., N. R. Waterfield, T. P. Beresford, R. W. Le Page, and A. W. Jarvis. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucchini, S., F. Desiere, and H. Brussow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macrina, F. L., R. P. Evans, J. A. Tobian, D. L. Hartley, D. B. Clewell, and K. R. Jones. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145-150. [DOI] [PubMed] [Google Scholar]
- 32.Maniloff, J., and H. W. Ackermann. 1998. Taxonomy of bacterial viruses: establishment of tailed virus genera and the order Caudovirales. Arch. Virol. 143:2051-2063. [DOI] [PubMed] [Google Scholar]
- 33.Masukata, H., and J. Tomizawa. 1986. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell 44:125-136. [DOI] [PubMed] [Google Scholar]
- 34.Masukata, H., and J. Tomizawa. 1990. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell 62:331-348. [DOI] [PubMed]
- 35.McKay, L. L., and K. A. Baldwin. 1984. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl. Environ. Microbiol. 47:68-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137:227-231. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pillidge, C. J., and A. W. Jarvis. 1988. DNA restriction maps and classification of the lactococcal bacteriophages c2 and sk1. New Zealand Dairy Sci. Technol. 23:411-416. [Google Scholar]
- 40.Powell, I. B., M. G. Achen, A. J. Hillier, and B. E. Davidson. 1988. A simple and rapid method for genetic transformation of lactic streptococci by electroporation. Appl. Environ. Microbiol. 54:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell, I. B., and B. E. Davidson. 1985. Characterization of streptococcal bacteriophage c6A. J. Gen. Virol. 66:2737-2741. [DOI] [PubMed] [Google Scholar]
- 42.Powell, I. B., and B. E. Davidson. 1986. Resistance to in vitro restriction of DNA from lactic streptococcal bacteriophage c6A. Appl. Environ. Microbiol. 51:1358-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell, I. B., D. L. Tulloch, A. J. Hillier, and B. E. Davidson. 1992. Phage DNA synthesis and host DNA degradation in the life cycle of Lactococcus lactis bacteriophage c6A. J. Gen. Microbiol. 138:945-950. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sandmeier, H. 1994. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibers. Mol. Microbiol. 12:343-350. [DOI] [PubMed] [Google Scholar]
- 46.Schouler, C., C. Bouet, P. Ritzenthaler, X. Drouet, and M. Mata. 1992. Characterization of Lactococcus lactis phage antigens. Appl. Environ. Microbiol. 58:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schouler, C., S. D. Ehrlich, and M. C. Chopin. 1994. Sequence and organization of the lactococcal prolate-headed bIL67 phage genome. Microbiology 140:3061-3069. [DOI] [PubMed] [Google Scholar]
- 48.Schouler, C., M. Gautier, S. D. Ehrlich, and M. C. Chopin. 1998. Combinational variation of restriction modification specificities in Lactococcus lactis. Mol. Microbiol. 28:169-178. [DOI] [PubMed] [Google Scholar]
- 49.Selzer, G., T. Som, T. Itoh, and J. Tomizawa. 1983. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell 32:119-129. [DOI] [PubMed] [Google Scholar]
- 50.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tetart, F., D. C., and H. M. Krisch. 1998. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: Recombination between conserved motifs swaps adhesin specificity. J. Mol. Biol. 282:543-556. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterfield, N. R., M. W. Lubbers, K. M. Polzin, R. W. Le Page, and A. W. Jarvis. 1996. An origin of DNA replication from Lactococcus lactis bacteriophage c2. Appl. Environ. Microbiol. 62:1452-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]