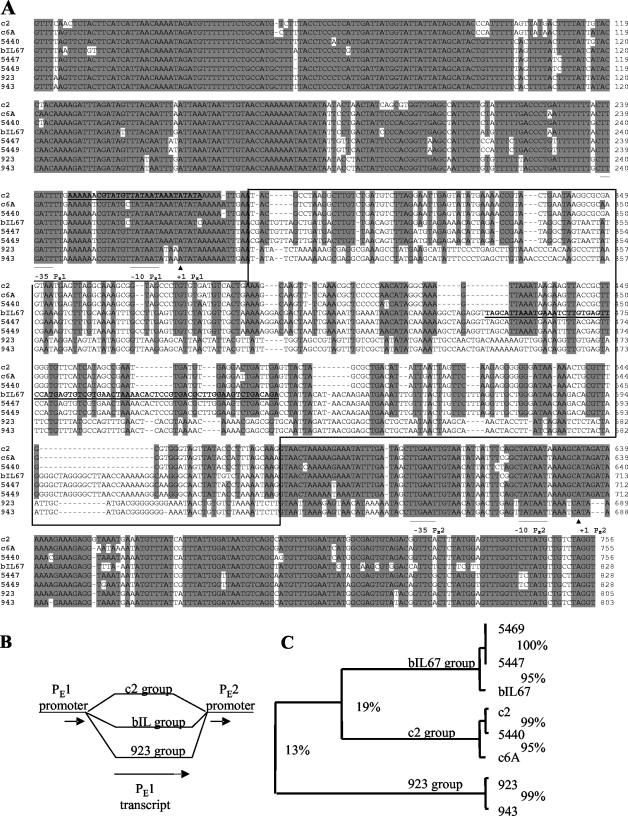
FIG. 1.
(A) Alignment of PE1 transcript template and flanking sequences. Sequences were aligned using the ClustalW multiple alignments program (52) within the BioEdit sequence analysis package (Tom Hall, North Carolina State University, Raleigh). The aligned sequences start at position 7294 and end at position 6540 of the c2 genomic sequence. The −35 and −10 boxes of PE1 and PE2 promoters are underlined, and the + 1 residues labeled with the symbol ▴ (28). The nonconserved portion of the alignment is boxed. The two highlighted sequences (in boldface type and underlined) showed significant identities with sequences from the NCBI nucleotide database: nt 247 to 275, 96% identity to the L. lactis IL1403 chromosome; nt 449 to 525, 88% identity to the small isometric phage bIL170; and nt 470 to 525 (contained within the preceding sequence), 92% identity to the plasmids pAW601 and pIL103. (B) Schematic representation of origin divergence. Single lines represent the conserved sequences and branched lines the divergent region; short arrows, promoters; long arrow, PE1 transcript template. Note that the c2 group PE1-T template is shorter than that of the bIL67 and 923 groups. The dotted portion of the arrow symbolizes the length difference. (C) Rectangular cladogram based on the ClustalW alignment of PE1 templates only (nonconserved portion of sequences, boxed in A). Numbers represent % identities between the phage and phage groups.