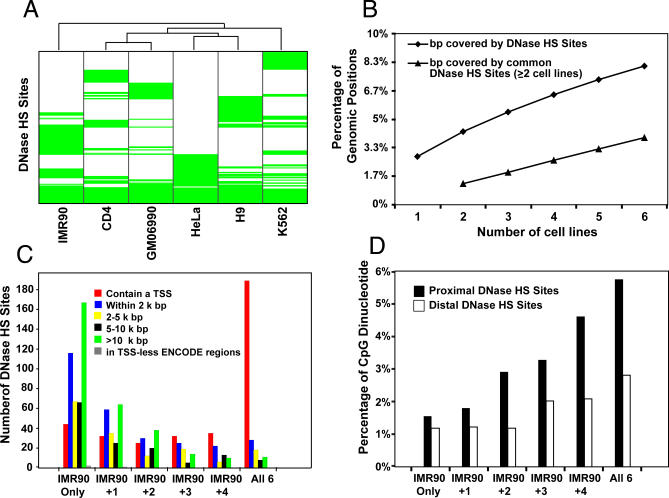
Figure 2. Identification and Characterization of DNaseI HS Sites.
(A) Clustering of six cell lines based on DNaseI HS site profiles. ENCODE regions were divided into 2-kb blocks and a binary DNaseI HS site profile was calculated for each cell line (1 for blocks containing a DNaseI HS sites hit, displayed in green, and 0 otherwise, uncolored). Cell lines were clustered based on their DNaseI HS sites hit profiles using Wards hierarchical clustering [41] with Euclidian distance as the metric. Ubiquitous DNase sites are grouped at the bottom.
(B) Cumulative percentage of the genome covered by DNaseI HS sites from increasing number of cell lines. Diamonds represent cumulative percentage of the genome covered by DNaseI HS sites from any cell line. Triangles represent cumulative percentage of the genome overlapped by DNaseI HS sites shared by at least two cell types. Each point is an averaged value of all possible cell line combinations.
(C) Location of DNaseI HS sites relative to TSS. DNaseI HS sites from IMR90 cells were first categorized as unique to IMR90, common with other cell types, or ubiquitous in all six cell types. Data centering on other cell types are identical (unpublished data). Distances of each DNaseI HS site were calculated to the nearest TSS.
(D) CpG dinucleotide distribution. The percentage of CG dinucleotide was determined for proximal and distal DNaseI HS sites that were unique to IMR90, common with additional cell types, or ubiquitous within all six cell types.