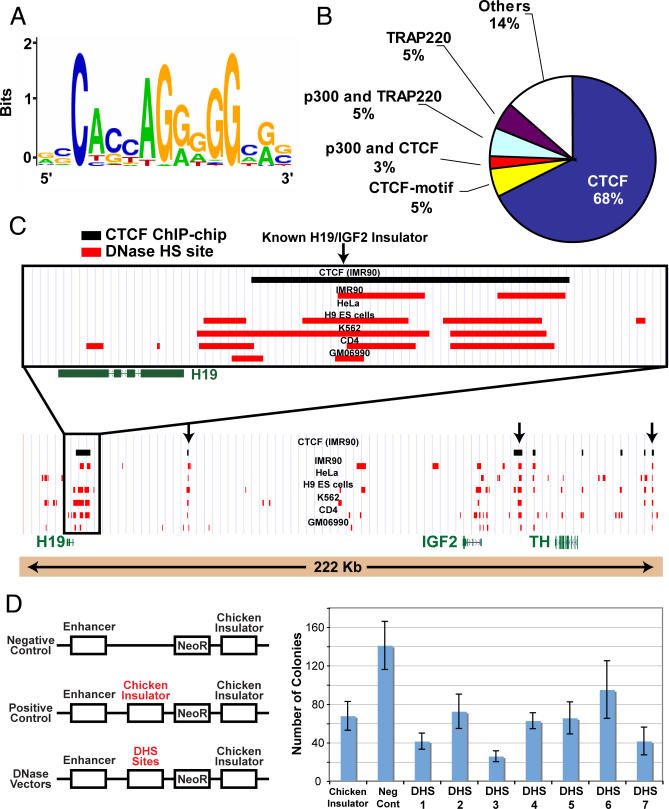
Figure 4. The CTCF Motif Is Identified in Ubiquitous Distal DNaseI HS Sites.
(A) Motif identified using de novo motif-finding algorithm using the minimal intersecting regions of ubiquitous distal DNaseI HS sites. The overall height of the stack of letters in a position indicates the sequence conservation at that position, while the height of letters within the stack indicates the relative frequency of each base at that position.
(B) Percentage of ubiquitous distal DNaseI HS sites that overlap with CTCF ChIP-chip data, CTCF motif, or other enhancer elements.
(C) Example of clustered ubiquitous DNaseI HS sites that overlap CTCF in the H19/IGF2 locus. Arrows indicate ubiquitous DNaseI HS sites overlapping with CTCF.
(D) Cell culture insulator assays demonstrate that DNaseI HS sites (that overlap CTCF) display enhancer-blocking activity. Higher colony counts in G418 media indicate no enhancer-blocking activity, while lower colony counts indicate positive enhancer-blocking activity. A previously described chicken insulator was used as a positive control [11].