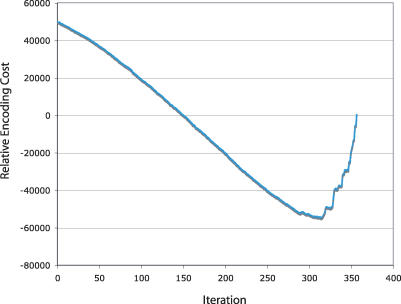
Figure 4. The Encoding Cost as a Function of the SCI-PHY Iteration for the Secretin Family.
We subtract the encoding cost of the null hypothesis (that all sequences belong in a single subfamily) from the cost of encoding the subclass alignments at each iteration of the algorithm (y-axis: Costiteration − Costnull). At program commencement, the number of subclasses equals the number of sequences and the encoding cost is high. The encoding cost curve decreases steadily to a minimum when similar sequences are joined and then increases as subtrees with different amino acid preferences are joined. The point in the agglomeration for which the encoding cost is minimal is used to determine a cut of the tree into subtrees, defining the SCI-PHY subfamily decomposition. If the minimum occurs when the encoding cost is zero, then all sequences are placed in a single class (i.e., no subfamilies are predicted). Negative “Encoding Cost” values indicate savings relative to the null hypothesis, and provide support for a division of the sequences into two or more subfamilies.