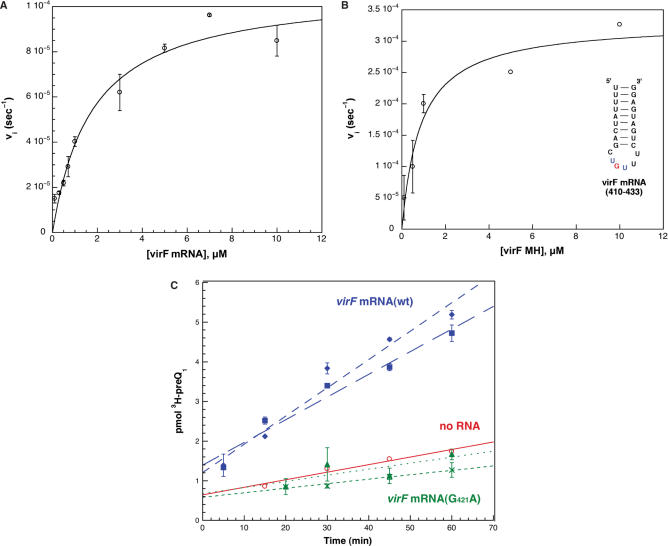
Figure 6.
Kinetic characterization of virF mRNA substrates with E. coli TGT and [3H] preQ1. (A) and (B) The individual data points were fit to the Michaelis–Menten equation by non-linear regression, and the average data points (with error bars representing the standard deviations of the averages) for each concentration of RNA substrate are depicted. The study was performed with duplicate kinetic trials at saturating concentrations of [3H] preQ1. The inset in B shows the structure of the RNA minihelix corresponding to bases 410–433 of the virF mRNA. (C) The comparison of radioactive substrate incorporated over time in virF mRNA(wt) (blue, squares and diamonds), virF mRNA(G421A) (green, crosses and triangles), and a no RNA control (red, circles), fit by linear regression. Each virF mRNA substrate was analyzed at two concentrations: 2 μM (squares and crosses) and 10 μM (diamonds and triangles). The average of duplicate trials is shown.