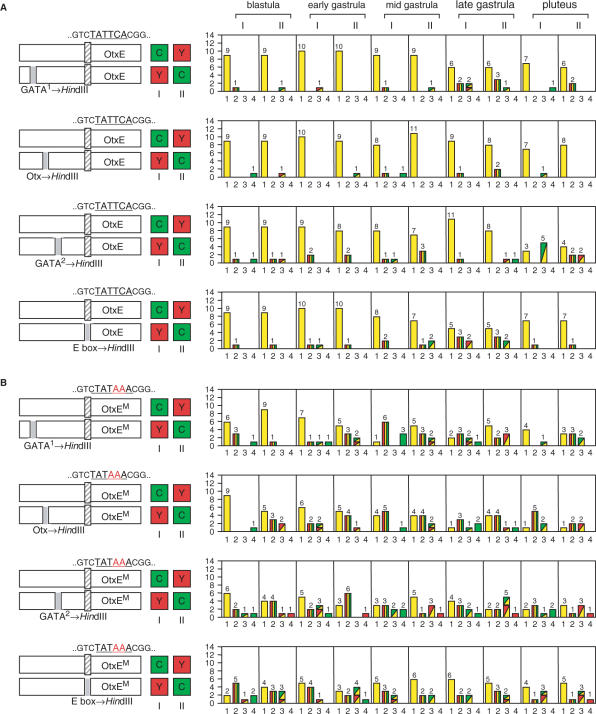
Figure 5.
The effect of the four putative transcription factor binding sites on spatiotemporal expression of the TATA-less and TATA-containing HpOtxE promoters. In each panel, the schematic on the left indicates the reporter plasmids introduced into fertilized eggs. I and II refer to the particular combinations of reporter (CFP or YFP) and promoter, as indicated by the boxes marked C and Y. The number of embryos with each expression pattern profile is summarized in the panel to the right of each schematic. (A) (First panel) The plasmids used were pM4705/pM4938 (I) and pM4937/pM4692 (II), driven by the HpOtxE promoter fused with the intact or mutated HpOtxE upstream sequence within the putative GATA factor binding site #1 (−360 to −355 bp). (Second panel) Similar analysis using pM4940 (YFP) and pM4939 (CFP), in which the HpOtxE upstream sequence was mutated within the putative Otx factor binding site (−294 to −289 bp), instead of pM4938 (YFP) and pM4937 (CFP) in the first panel. (Third panel) Analysis using pM4942 (YFP) and pM4941 (CFP), in which the HpOtxE upstream sequence was mutated within the putative GATA factor binding site #2 (−250 to −245 bp). (Fourth panel) Analysis using pM4783 (YFP) and pM4782 (CFP), in which the HpOtxE upstream sequence was mutated within the E-box (−66 to −61 bp). (B) (First panel) pM4694/pM7065 (I) or pM7062/pM4706 (II), which are driven by the mutated HpOtxE core promoter (OtxEM as described in Figure 3A) fused with the intact or mutated HpOtxE upstream sequence within the putative GATA factor binding site #1. (Second panel) pM7066 (YFP) and pM7063 (CFP), in which the HpOtxE upstream sequence was mutated within the putative Otx factor binding site. (Third panel) pM7067 (YFP) and pM7064 (CFP), in which the HpOtxE upstream sequence was mutated within the putative GATA factor binding site #2. (Fourth panel) pM4781 (YFP) and pM4780 (CFP), the HpOtxE upstream sequence was mutated within the E-box.