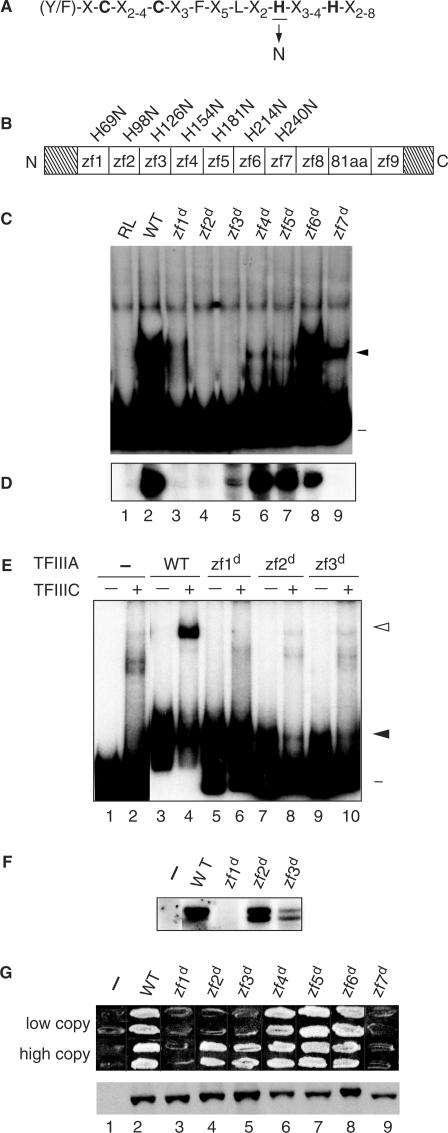
Figure 1.
Zinc finger 1 and zinc finger 7 are essential for the TF activity of TFIIIA. (A) The consensus amino acid sequence of the Cys2His2 zinc finger is given in single-letter code and the zinc-coordinating cysteines and histidines are in bold. The histidine substituted with asparagine to disrupt the structure of individual fingers is underlined. (B) Schematic representation of yeast TFIIIA. The boxes denoted zf1 to zf9 represent the nine zinc fingers; the boxed 81aa denotes the 81-amino-acid domain; and the diagonally stripped boxes represent the 48-amino-acid amino-terminal and the 35-amino-acid carboxyl-terminal regions. The histidine-to-asparagine substitution for each disruption is denoted above the zinc finger. zf = zinc finger. (C) Ability of mutant versions of TFIIIA to bind to the 5 S RNA gene as assessed by EMSA. A radioactively labeled DNA fragment containing the yeast 5 S RNA gene was incubated with in vitro synthesized versions of TFIIIA prior to electrophoresis on a non-denaturing polyacrylamide gel. Lane 1, RL (reticulocyte lysate; in vitro transcription–translation reaction that was not programmed to synthesize TFIIIA); lane 2, WT (wild-type) TFIIIA; lanes 3 through 9, versions of TFIIIA containing a histidine-to-asparagine mutation in the indicated zinc finger. Lane numbers are as given in panel D. The positions of the free DNA (minus sign) and the TFIIIA–DNA complex (closed arrowhead) are indicated on the right. (D) Abilities of mutant versions of TFIIIA to support in vitro transcription of the 5 S RNA gene. In vitro transcription reaction mixtures contained the yeast 5 S RNA gene as template, partially purified yeast TFIIIC, TFIIIB and RNA polymerase III and the version of in vitro synthesized TFIIIA indicated in the corresponding lanes of panel C. The RNAs synthesized in vitro were analyzed on a 7 M urea–10% polyacrylamide gel. The autoradiogram shows the portion of the gel containing 5 S RNA. (E) Abilities of mutant versions of TFIIIA purified from bacteria to bind to the 5 S RNA gene and to recruit TFIIIC to the TFIIIA–DNA complex as assessed by EMSA. A radioactively labeled DNA fragment containing the yeast 5 S RNA gene was incubated with protein extract containing the indicated version of bacterially produced TFIIIA in the absence (odd numbered lanes) or presence (even numbered lanes) of partially purified yeast TFIIIC prior to electrophoresis on a non-denaturing polyacrylamide gel. Lanes are labeled as in panel C with lanes 1 and 2 containing no added bacterial protein. The positions of the free DNA (minus sign), TFIIIA–DNA complexes (solid arrowhead) and TFIIIC–TFIIIA–DNA complexes (arrowhead) are indicated at the right. (F) Abilities of mutant versions of TFIIIA purified from bacteria to support in vitro transcription of the 5 S RNA gene. For details, see legend for Figure 1D. (G) Abilities of mutant versions of TFIIIA to support cell viability. Top panel: a plasmid shuffle assay to test the abilities of mutant versions of TFIIIA to replace wild-type TFIIIA in vivo. Cells of YRW1 that had been transformed with plasmids containing copies of the TFC2 gene were tested for their abilities to grow on medium containing 5-FOA (see Results Section). Each patch contains cells from a different transformant. Two series of plasmids were tested: in the pRS314+ series (labeled ‘low copy’), TFC2 is expressed under the control of its own promoter from a low-copy (CEN ARS) plasmid; in the ΔpG3 series (labeled ‘high copy’), TFC2 is expressed untrol of the strong GPD promoter from a high-copy (2 μ) plasmid. Cells containing the parental vectors not encoding a version of TFIIIA are indicated by the minus sign. Bottom panel: assessment by western blot of in vivo protein levels of mutant versions of TFIIIA. Aliquots of crude lysates prepared from representative YRW1 yeast cells containing the indicated ΔpG3-derived plasmids were separated on a 15% SDS polyacrylamide gel, transferred to a nitrocellulose filter, and probed with anti-TFIIIA antibody. Note that TFIIIA expressed from the pRS314+ series of plasmids is below the level of detection in this blot.