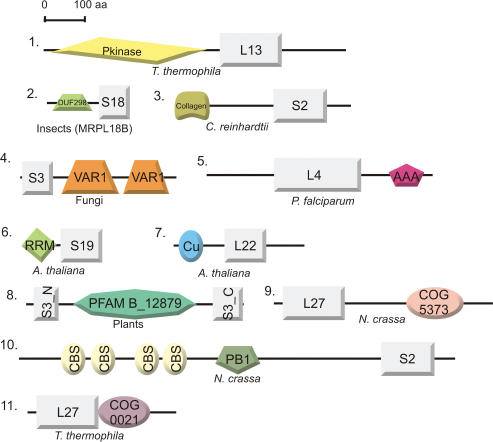
Figure 7.
Domain architectures of MRPs to which additional functional domains have been added. Protein identifiers are depicted as in Table 1 or 2 and species distribution is indicated. Proteins are drawn approximately according to scale. Domain abbreviations: (1) Pkinase (pfam00069): Protein kinase catalytic domain. (2) DUF298 (pfam03556): Domain of unknown function, containing a basic helix-loop-helix leucine zipper motif. (3) Collagen (pfam01391): Collagen triple helix repeat. (4) VAR1 (pfam05316): This domain is specific for some fungal mitochondrial ribosomal S3 proteins and is often present in two copies (also see text). (5) AAA (pfam00004): ATPase family associated with various cellular activities. Proteins containing AAA domains often perform chaperone-like functions that assist in the assembly, operation or disassembly of protein complexes. (6) RRM (pfam00076): RNA-binding domain, found in a variety of RNA-binding proteins (also see text). (7) Cu (pfam02298): Copper-binding domain found in various proteins (also see text). (8) In plants, the N-terminus (S3_N, pfam00417) and C-terminus (S3_C, pfam00189) of MRPS3 are separated as a result of an insertion of a protein domain of unknown function (pfam B domain PB012879). (9) COG5373: Predicted membrane protein present in prokaryotes. (10) CBS (pfam00571) and PB1 (pfam00564) domains: CBS domains are small intracellular modules, which have a regulatory role in making proteins sensitive to adenosyl carrying ligands. PB1 is present in many eukaryotic cytoplasmic signaling proteins. (11) COG0021: Transketolase (1-deoxyxylulose-5-phosphate synthase).