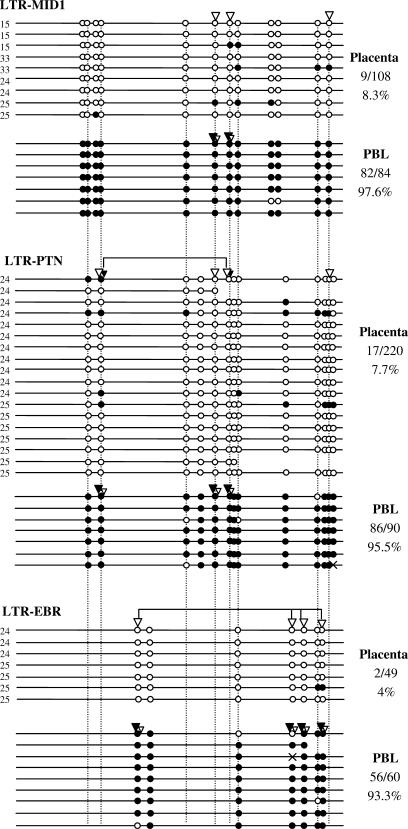
Figure 2.
Methylation of HERV-E LTR-derived gene promoters. Each line corresponds to a unique sequence of an LTR. Black circles correspond to methylated CpGs and white circles to unmethylated CpGs. Homologous CpGs between LTRs are connected by dashed lines. White and black triangles indicate unmethylated and methylated CpG sites, respectively, revealed by COBRA. Black and white triangles present at the same CpG site indicate that the population of amplified sequences contained both methylated and unmethylated CpGs at this site. In this case, the triangle's relative size corresponds to the relative band intensity of the digested and undigested forms. Connectors indicate CpG sites that were assayed by the same restriction enzyme in COBRA (see Materials and Methods section). Crosses correspond to mutated CpG sites. The number of methylated CpGs out of the total CpG sites is reported for each LTR. Numbers 24, 25, etc. on the left of each sequence indicate the corresponding placenta sample (P24, P25, etc).