Fig. 4.
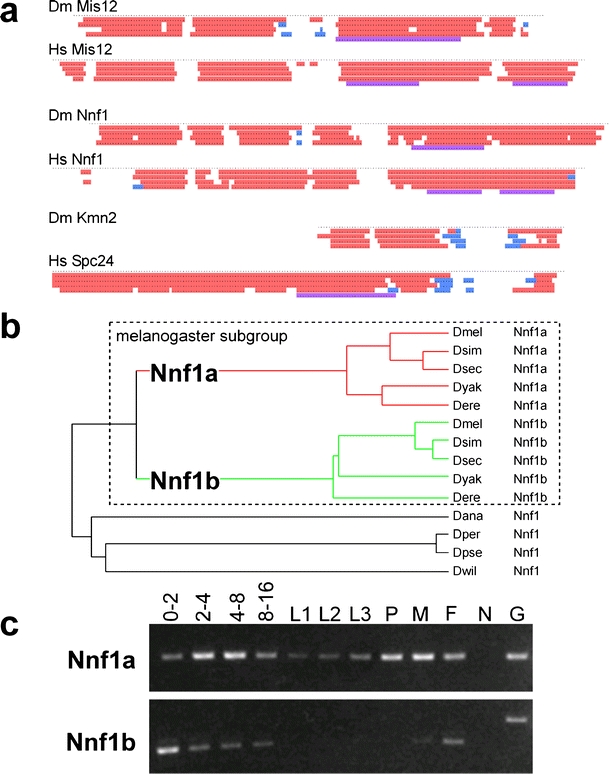
Secondary structure and genomic comparison of Drosophila kinetochore proteins. a Secondary structure predictions reveal similarities between Drosophila (Dm) and human (Hs) homologs of Mis12, Nnf1, and between Drosophila Kmn2 and the C-terminal region of human Spc24. Each dot represents an amino acid position. The lines with colored regions illustrate predictions obtained with various algorithms (PSIPRED, JNET, PROF Quali and King, PROF Rost, COILS from top to bottom). α-helical regions are shown in red, regions with β-sheets in blue and coiled coils in magenta. b A phylogenetic tree was constructed after aligning the predicted amino acids sequences encoded by Nnf1-like genes in Drosophilid genomes. An Nnf1 gene duplication resulting in the two paralogs Nnf1a and Nnf1b early after the divergence of the melanogaster subgroup lineage provides the most parsimonious explanation for the observed branching pattern. D. melanogaster (Dmel), D. simulans (Dsim), D. sechellia (Dsec), D. yakuba (Dyak), D. erecta (Dere), D. ananassae (Dana), D. persimilis (Dper), D. pseudoobscura (Dpse), D. willistoni (Dwil). Synteny considerations (data not shown) indicate that Nnf1a represents the primordial homolog. c The developmental expression pattern of D. melanogaster Nnf1a and Nnf1b was analyzed by RT-PCR experiments. The stages analyzed were: embryos 0–2 (0–2), 2–4 (2–4), 4–8 (4–8), and 8–16 (8–16) hours after egg deposition; larval stages (L1, L2, L3), pupae (P), adult males (M) and females (F). Control amplifications (G) from a cloned Nnf1a cDNA and an intron containing genomic Nnf1b fragment, as well as amplifications (N) with mock reverse transcribed mRNA demonstrated that the RT-PCR products were not derived from contaminating genomic DNA. The results are consistent with Nnf1a expression being correlated with mitotic proliferation and Nnf1b expression being germline-specific
