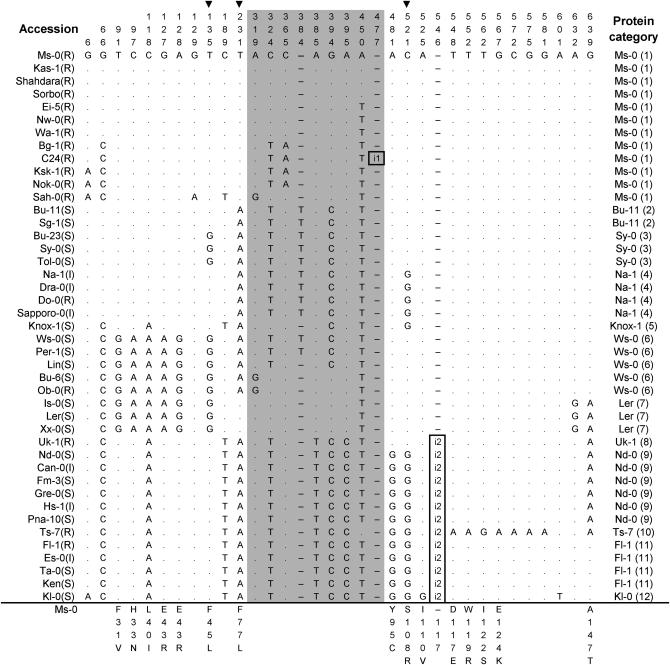
Figure 3.—
Polymorphic sites of RPW8.1 aligned against the Ms-0 RPW8.1 allele (GenBank accession no. AF273059). A dot indicates an identical nucleotide and a dash indicates a gap. Shaded are the substitutions in the intron. The numbers at the top indicate the nucleotide position relative to the start codon of the Ms-0 alleles. Amino acid replacements caused by nucleotide substitutions are indicated at the bottom. The arrowhead at the top indicates the nonsynonymous substitution that may affect functionality of the proteins (site that showed statistically significant association with the DR phenotype at the 5% level after Bonferroni correction). The disease phenotypes (R, resistant; I, intermediate; S, susceptible) are indicated in the parentheses after the names of the accessions. Identical deduced proteins are represented by one allele only and the numbers in the parentheses indicate the group numbers. Boxed are two insertions: i1, an insertion of 10 bp (GTTTATCTTT); and i2, an insertion of 63 bp (GATCAATGGGACGATATCAAAGAAATCAAGGCCAAGATATCTGAAACGGACACTAAACTTGCT), which was an intragenic duplication (from nucleotide 546–608) and resulted in an insertion of 21 amino acids (DQWDDIKEIKAKISETDTKLA).