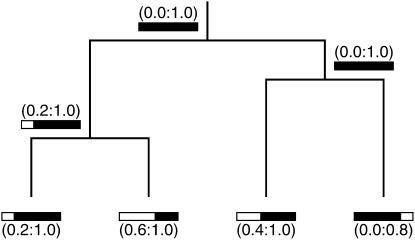
Figure 3.—
Illustration of how g is determined for both extant and ancestral sequences as one works backward in time toward their MRCA. The history for four sampled sequences, each with missing data, is shown with their known and unknown regions represented as solid and open bars. The full-length alignment for these four extant sequences is 1000 bases. The first number for each sampled sequence marks the relative alignment position at the end of its unknown region (if any) to its left. For example, the leftmost sampled sequence is missing the first 200 bases of the full-length alignment. In turn, the following interval in parentheses corresponds to its g [i.e., its summary of known regions as scored over the closed interval of (0:1) for the full alignment]. Thus, g = (0.2:1.0) for the first extant sequence. As one works backward in time and coalescent events are accounted for, g is then calculated for each ancestral sequence as the union of g for its two coalescing sequences [e.g., g = (0.2:1.0) for the common ancestor of the two leftmost sampled sequences]. In these ways, known and unknown regions of both extant and ancestral sequences are tracked back to the MRCA of the population sample.